CRISPR/Cas9 system suitable for saccharomyces polygene knockout as well as construction method and application of CRISPR/Cas9 system
A construction method and gene knockout technology, applied in the field of genetic engineering, can solve the problem of low efficiency of multi-gene knockout, and achieve the effect of high knockout efficiency, low cost, and promotion of genetic engineering transformation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
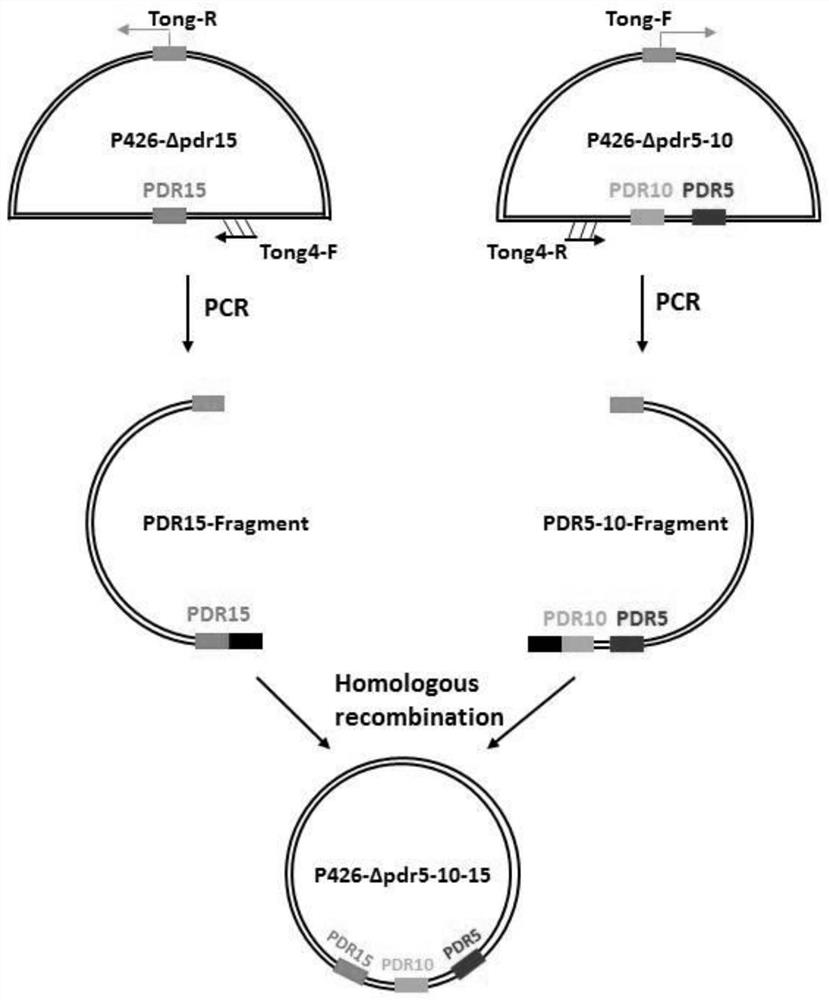
[0036] Example 1: Construction of Saccharomyces cerevisiae BJ5464 multiple gene knockout vector
[0037] 1. Selection of knockout targets
[0038] (1) First search and obtain the pdr5 (Gene ID: 854324), pdr10 (GeneID: 854506), pdr15 (GeneID: 854506) and pdr15 (GeneID: 852015) gene sequence.
[0039](2) Using the CRISPR tool (http: / / crispr.dbcls.jp / ), upload the target sequence and analyze it to obtain all potential target sites of the target gene, that is, all downstream 20bp sequences containing NGG characteristic bases. The sequence with higher evaluation score and closer to the start codon ATG was selected as the gRNA knockout target.
[0040] 2. Construction of single knockout vector
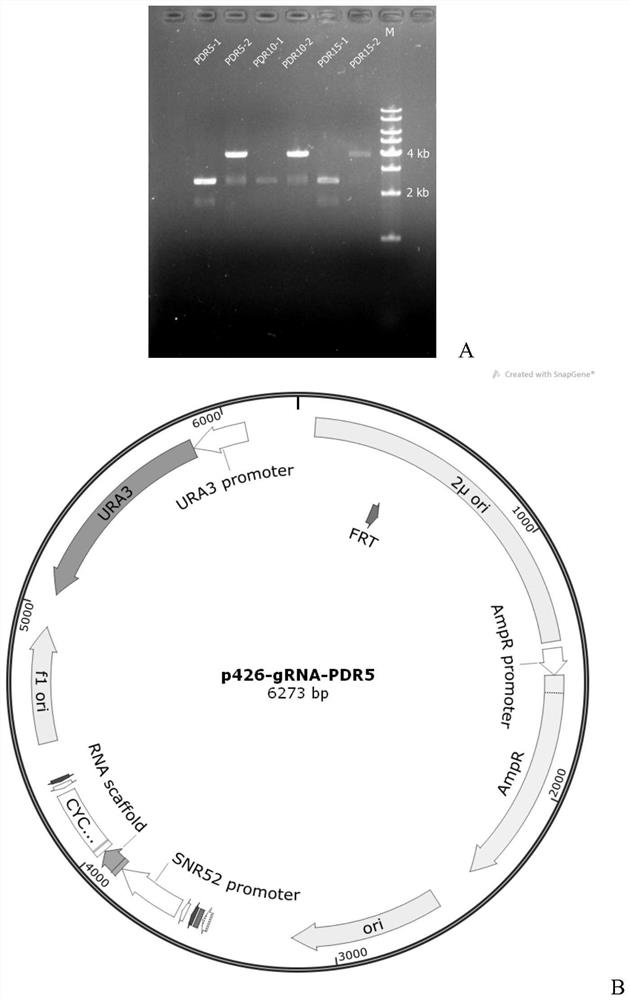
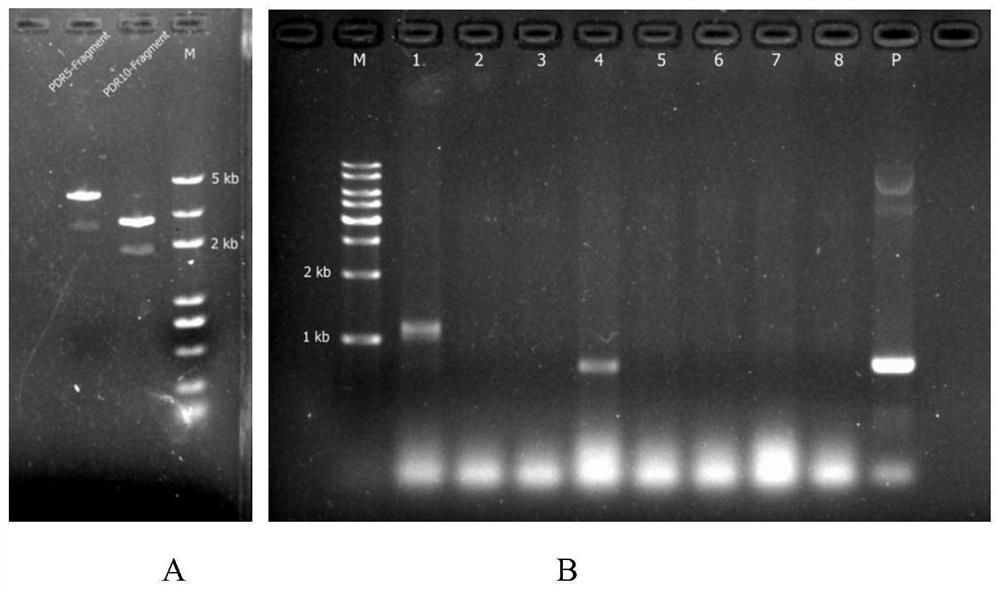
[0041] Based on the CRISPR knockout vector p426-PSNR52-gRNA.CAN1.Y-TSUP4, three single gene knockout plasmids (single knockout plasmids): p426-Δpdr5, p426-Δpdr10 and p426-Δpdr15 were constructed. Taking the construction of the vector p426-Δpdr5 targeting the target gene pdr5 as an exampl...
Embodiment 2
[0078] Example 2: Knockout of S. cerevisiae BJ5464 toxin resistance gene:
[0079] 2.1 Preparation of yeast competent cells
[0080] The yeast transformation kit S.c.EasyComp Transformation Kit (Invitrogen) was used to prepare competent cells of the yeast strain S.cerevisiae BJ5464, and the specific method was as follows:
[0081] (1) Pick a single colony of yeast and inoculate it in 5mL YPD medium, culture it overnight at 30°C and 220rpm until the OD600 value is between 3.0-5.0;
[0082] (2) Dilute the bacteria: transfer the bacteria solution into 5mL YPD medium, make the initial OD600 value between 0.2-0.4, cultivate it at 30°C, 220rpm for about 4-6 hours, and its OD 600 The value reaches 0.6-1.0;
[0083] (3) Collect bacterial cells: put 5 mL of bacterial liquid in a 50 mL centrifuge tube, centrifuge at 4000 g for 5 min at room temperature, and discard the supernatant;
[0084] (4) Wash away the residual medium: Take 5mL Solution I (Wash solution) to resuspend the bacter...
Embodiment 3
[0092] Example 3: Research on the ability of Saccharomyces cerevisiae to resist exogenously added toxins
[0093] After knocking out the pdr5, pdr10 and pdr15 genes of Saccharomyces cerevisiae and obtaining the toxin-sensitive strain BJ5464-D-, it is necessary to investigate the tolerance of the strain to the addition of exogenous toxin DON. There are two main purposes: (1) verification After knocking out the three genes of the pdr series, does it reduce or weaken the ability of Saccharomyces cerevisiae to resist exogenous toxins? (2) To lay the foundation for selecting the appropriate toxin concentration and verifying the function of suspected anti-toxin genes in the later stage. Therefore, this study investigated the tolerance of Saccharomyces cerevisiae to DON before and after knockout.
[0094] On the YPD plate with 100 μM DON toxin concentration, the Saccharomyces cerevisiae colony was obviously sparse and blurred with the increase of the dilution gradient, and the OD va...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com