Method for identifying and detecting porphyra red rot pythium pathogens
A technology for identifying detection and detection methods, which is applied in the directions of microorganism-based methods, biochemical equipment and methods, and microorganism determination/inspection. , the effect of shortening the detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0034] The present invention will be described in detail below in conjunction with the accompanying drawings and examples.
[0035] 1. Strains
[0036] The strains used in this example come from the Japan Biological Resources Bank (NITE), the China General Microbiological Culture Collection Center (CGMCC) and the strains isolated and preserved in our laboratory. See Table 1 for details. . Among them, P.porphyrae and P.chondricola were grown at 25°C in semi-seawater corn agar medium; other strains of the genus Pythium and Alternaria were grown at 25°C in PDA medium (Haibo Biology, Qingdao); Pseudoalteromonas carrageenans and Bacillus huatsunata grew in Zobell 2216 medium at 20°C.
[0037] Bacterial strains used in the present embodiment of table 1
[0038]
[0039]
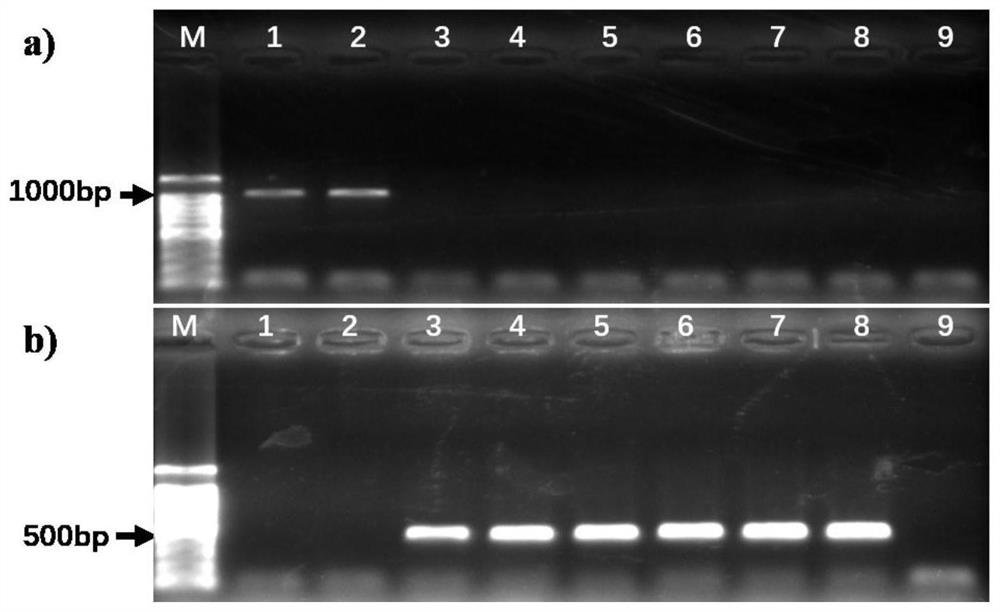
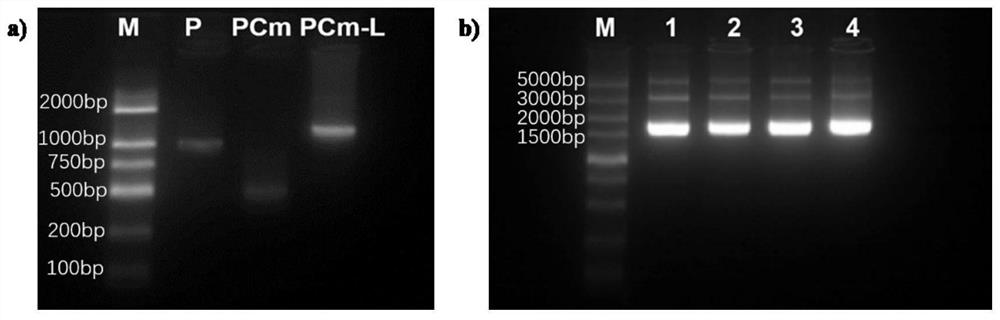
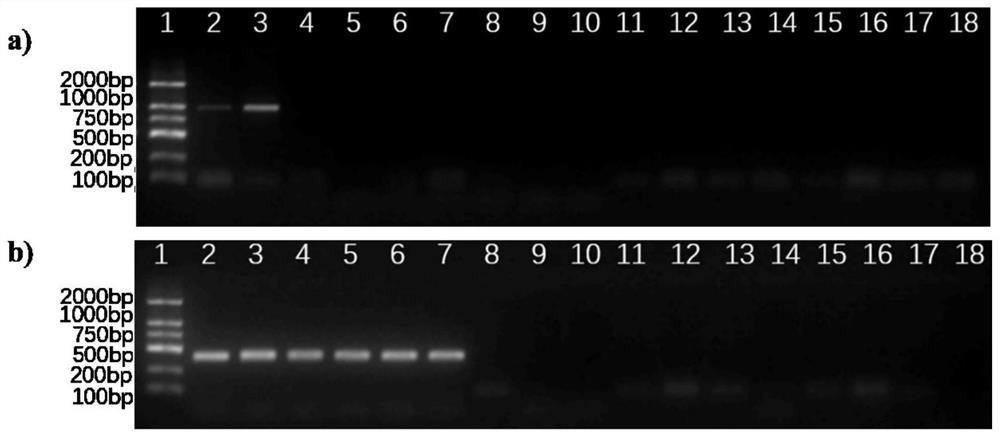
[0040] 2. Primer design and optimization of single-plex PCR conditions
[0041] 1. Primer Design
[0042] Genomic DNA of pure cultured P.porphyrae NBRC NO.30800 and P.chondricola NBRC NO.33253 hyphae was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com