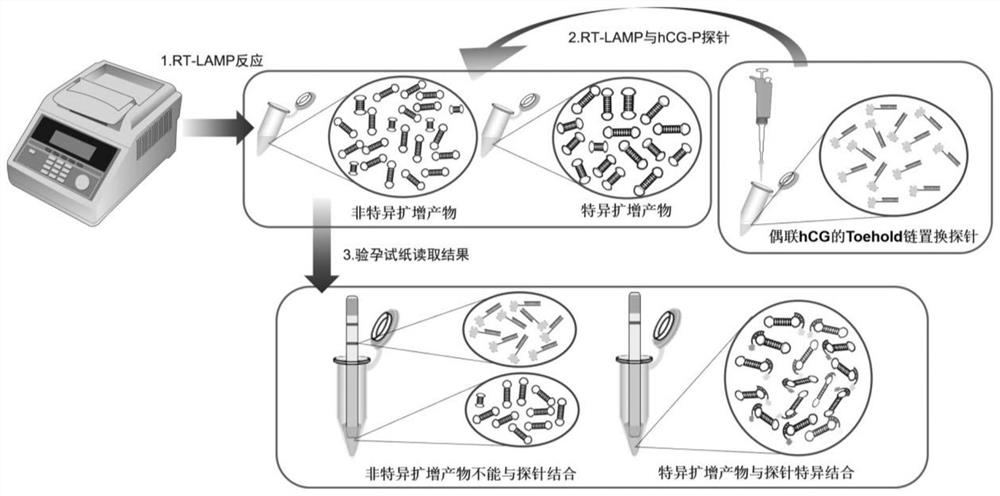
Primer, probe and kit for detecting neocoronavirus SARS-CoV-2
A sars-cov-2 and kit technology, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problem of low detection accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0060] The preparation method of the above-mentioned M-hCG-P1 comprises the following steps:
[0061] Step 1, modifying the M-P1 sequence to BG to prepare the M-BG-P1 sequence
[0062] Mix the M-P1 probe, BG-GLA-NHS, and HEPES in the solvent, incubate with shaking at room temperature for 2-2.5 hours, and separate on a desalting column to obtain the M-BG-P1 sequence.
[0063] The substance ratio of M-P1 probe, BG-GLA-NHS, and HEPES is 1:30:200;
[0064] Step 2: Preparation of hCG-SNAP fusion protein
[0065] HEK293T cells were transfected with pcDNA3.4-hCG-SNAP-His using PEI transfection reagent, and the cell supernatant was collected after 48 hours of culture, and the cell supernatant was filtered with a 0.45 μm filter, separated by an affinity chromatography column, and dialyzed to obtain hCG-SNAP fusion protein;
[0066] Step 3: Complete the connection between M-BG-P1 and hCG-SNAP fusion protein
[0067] Mix the hCG-SNAP fusion protein with the M-BG-P1 probe according to...
Embodiment 1
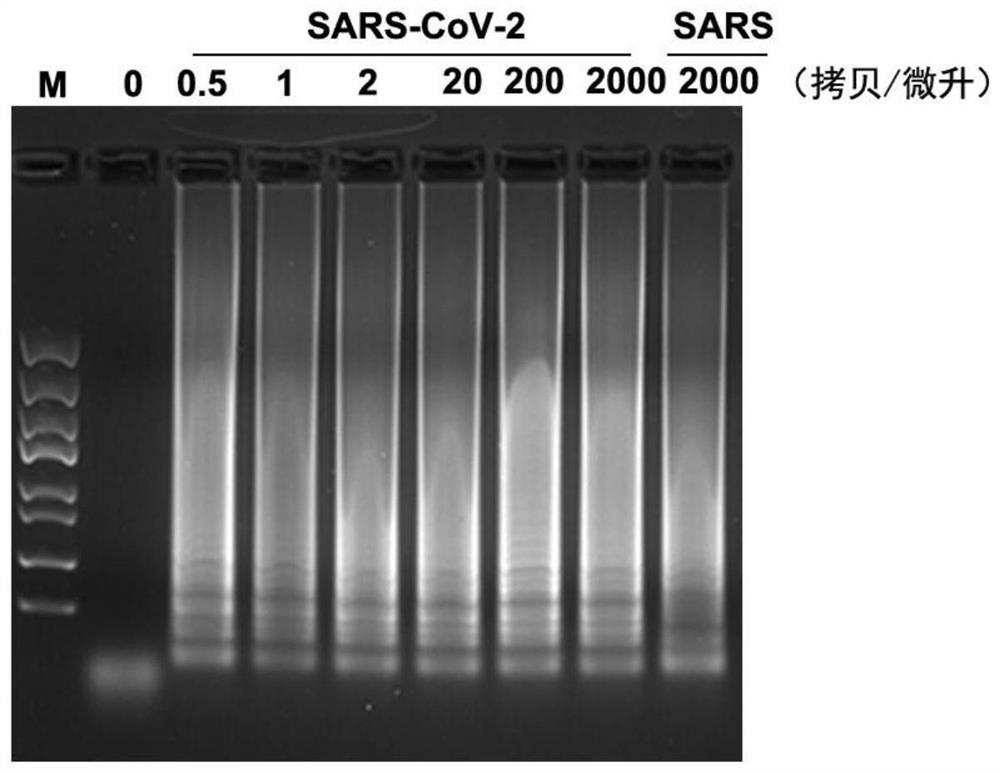
[0094] The M gene primers used to detect the new coronavirus SARS-CoV-2 are M-F3, M-B3, M-FIP, M-BIP and M-LP according to the material ratio of 1:1:4:4:2 wherein, the sequence of M-F3 is TAGGCTTGATGTGGCTCA; the sequence of M-B3 is AAGATGTCCACGAAGGATC; the sequence of M-FIP is CTGGATTGAATGACCACATGGAAGCTACTTCATTGCTTCTTTCAG; the sequence of M-BIP is ACATTCTTCTCAACGTGCCACACAGCTCCGATTACGAGTT; the sequence of M-LP is CGCGTACGCGCAAACAGT.
[0095] Amplify the M gene sequence of SARS-CoV-2 new coronavirus based on RT-LAMP method: use 25 μL RT-LAMP reaction system: 1×Isothermol Buffer contains 4 μL of the analyte, 0.6 μM M-B3, 0.6 μM M-F3, 2.4 μM M-FIP, 2.4 μM M-BIP, 1.2 μM M-LP, 1.12 mM dNTPs, 2MmMgSO 4 , 1mM betaine, 12UBst 2.0polymerase, 7.5UwarmstartRTx Reverse Transcriptase (when the pUC57-M plasmid is used as the test object, this reverse transcriptase is not added). The reaction conditions are as follows: first react at 58°C for 1.5h, then react at 80°C for 20min. The reaction...
Embodiment 2
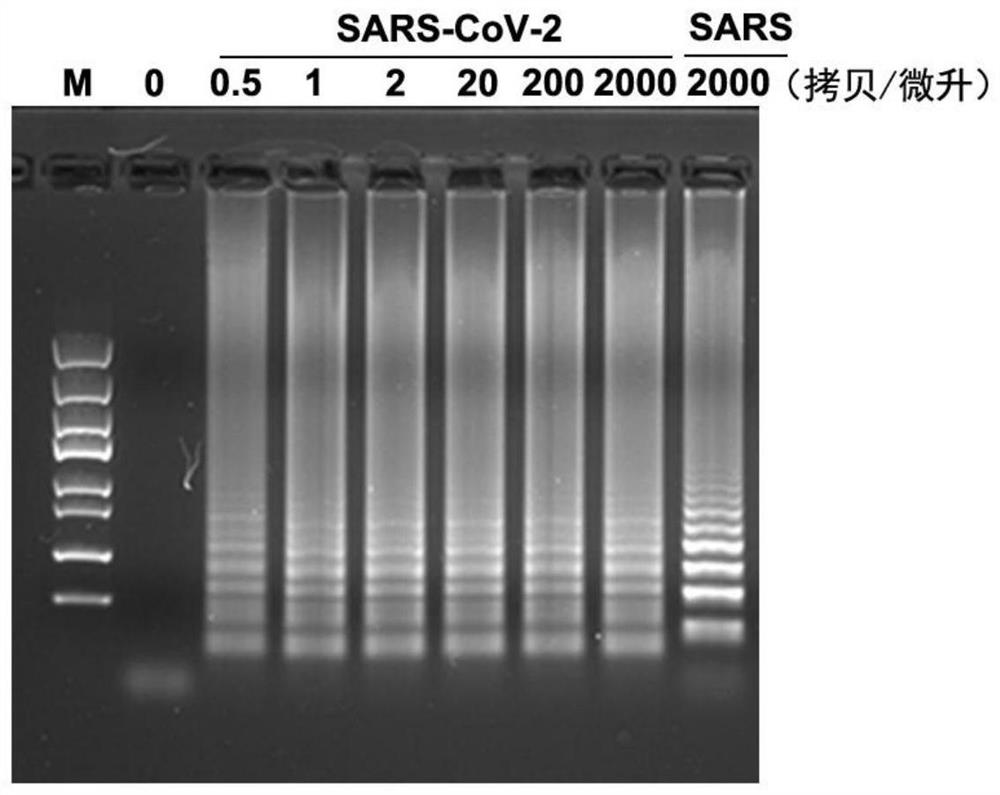
[0101] The N gene primers used to detect the new coronavirus SARS-CoV-2 are N-F3, N-B3, N-FIP, N-BIP and N-LP according to the ratio of the substance to 1:1:4:4:2 wherein, the sequence of N-F3 is TTGGCTACTACCGAAGAGCT; the sequence of N-B3 is TGCAGCATTGTTAGCAGGATT; the sequence of N-FIP is CTGGCCCAGTTCCTAGGTAGTACAGACGAATTCGTGGTGGTG; the sequence of N-BIP is GACGGCATCATATGGGTTGCAAGCGGGTGCCAATGTGAT;
[0102] Amplify the N gene sequence of SARS-CoV-2 new coronavirus based on RT-LAMP method: use 25 μL RT-LAMP reaction system: 1×Isothermol Buffer contains 4 μL of the analyte, 0.6 μM N-B3, 0.6 μM N-F3, 2.4 μM N-FIP, 2.4 μM N-BIP, 1.2 μM N-LP, 1.12 mM dNTPs, 4 mM MgSO 4 , 0.6mM betaine, 12U Bst 2.0polymerase, 3.75U WarmstartRTx Reverse Transcriptase (when the pUC57-N plasmid is used as the test object, this reverse transcriptase is not added). The reaction conditions are as follows: first react at 58°C for 1.5h, and then react at 80°C for 20min. The reaction device adopts an isother...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com