Double-stranded nucleic acid cyclization method, methylation sequencing library construction method and kit
A double-stranded nucleic acid and construction method technology, applied in the field of sequencing, can solve the problems of high methylation rate and low efficiency, and achieve the effect of reducing the high methylation rate
Pending Publication Date: 2021-05-14
BGI GENOMICS CO LTD
View PDF0 Cites 11 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
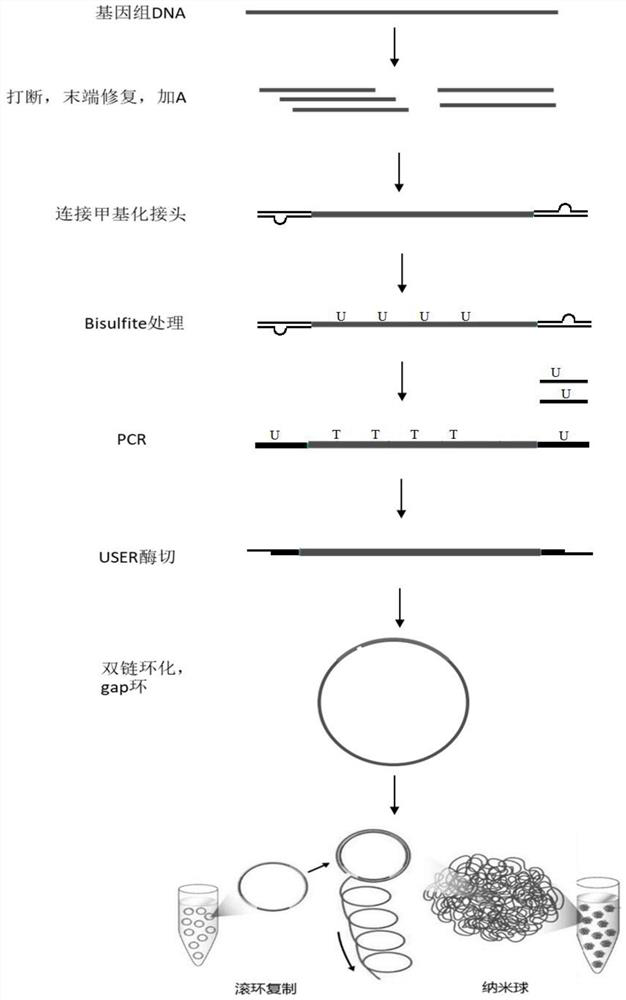
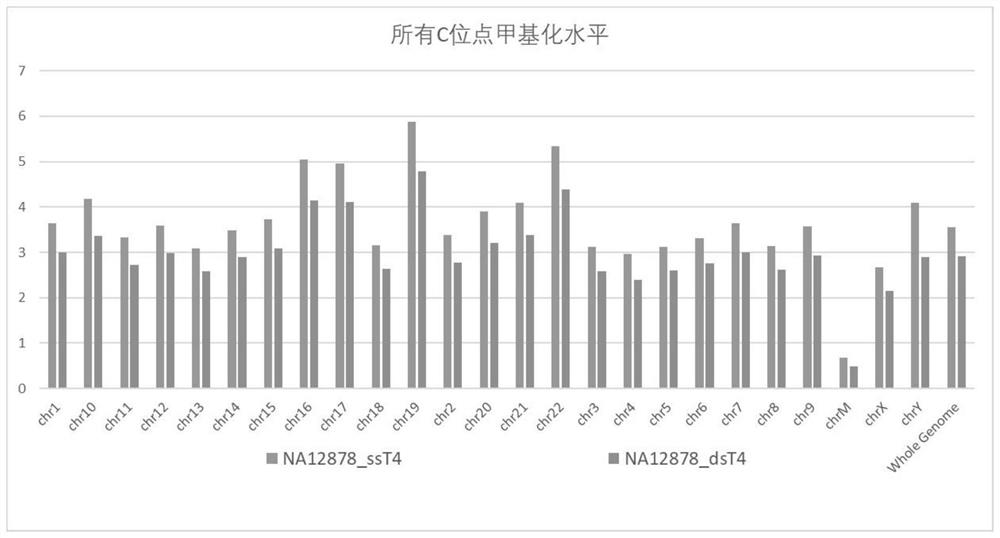
The efficiency of the circularization process is low, not all single-stranded DNA can self-link into a circle, and the shape of single-stranded DNA is more likely to form a secondary structure, resulting in the bias of the final circularization product, especially for the methyl group Whole genome bisulfite sequencing (WGBS), a library rich in AT bases, has a more prominent bias, and ultimately shows a high methylation rate
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment
[0079] In this example, two sets of libraries were constructed.
[0080] The control group adopts the method of commercialized kit (1000005251-MGIEasy Whole Genome Methylation Library Preparation Kit), that is, PCR primers without U base and single-strand circularization to carry out genome-wide methylation of human standard DNA (NA12878). Kylated WGBS library construction.
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
The invention discloses a nucleic acid double-chain cyclization method, a methylation sequencing library construction method and a kit. The nucleic acid double-strand cyclization method comprises the following steps: providing a forward primer and a reverse primer, wherein each of the forward primer and the reverse primer comprises a U basic group, the nucleotide number difference of the upstream sequences of the 5'ends of the U basic groups of the forward primer and the reverse primer is N, and the upstream sequences of the 5 'ends of the U basic groups of the forward primer and the reverse primer are at least partially and reversely complementary; amplifying the nucleic acid fragment by using the primer pair to obtain an amplification product; a USER enzyme is used for cutting off U basic groups in the amplification product, then a mononucleotide gap is formed, chain dissociation occurs to the residual nucleotide sequence from the gap to the 5'end due to the unstable structure, and then partially complementary cohesive terminal structures are formed at the two ends of the amplification product; the sticky tail end structures at the two ends are complementarily connected to form a double-chain structure, one chain of the double-chain structure is closed, and the other chain of the double-chain structure has N deleted basic groups. The method is used for constructing the WGBS library, and the phenomenon that the methylation rate of the WGBS library is high due to single-chain cyclization deviation can be reduced.
Description
technical field [0001] The invention relates to the technical field of sequencing, in particular to a double-stranded nucleic acid circularization method, a methylation sequencing library construction method and a kit. Background technique [0002] As an open platform, BGISEQ-500 and MGI2000, the sequencing platform of BGI, provide a one-stop sequencing operation process with a clear and clear sequencing process. It not only provides optional library preparation system and sample loading system, but also supports a variety of supporting library preparation The solution, using optimized combined probe-anchored aggregation technology (cPAS) and improved DNA nanoball (DNB) core sequencing technology, is one of the industry-leading high-throughput sequencing platforms. [0003] When constructing the sequencing library corresponding to this type of platform, the PCR library needs to be denatured into a single strand, and the single strand DNA is self-circularized with the help of...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Applications(China)
IPC IPC(8): C12Q1/6806C12N15/10C40B50/06
CPCC12Q1/6806C12N15/1093C40B50/06C12Q2531/113C12Q2523/125C12Q2531/125C12Q2521/531
Inventor 唐冲王娟何长寿童益琴杨林峰高强
Owner BGI GENOMICS CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com