High-flux visible chip detecting method of protein-protein interaction and detecting kit for protein-protein interaction
A protein interaction and chip detection technology, applied in the biological field, can solve the problems of limiting the number of testing samples, cultivating a large number of cells, prolonging the experimental process, etc., and achieve the effect of reducing experimental cost, simplifying the experimental process, and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
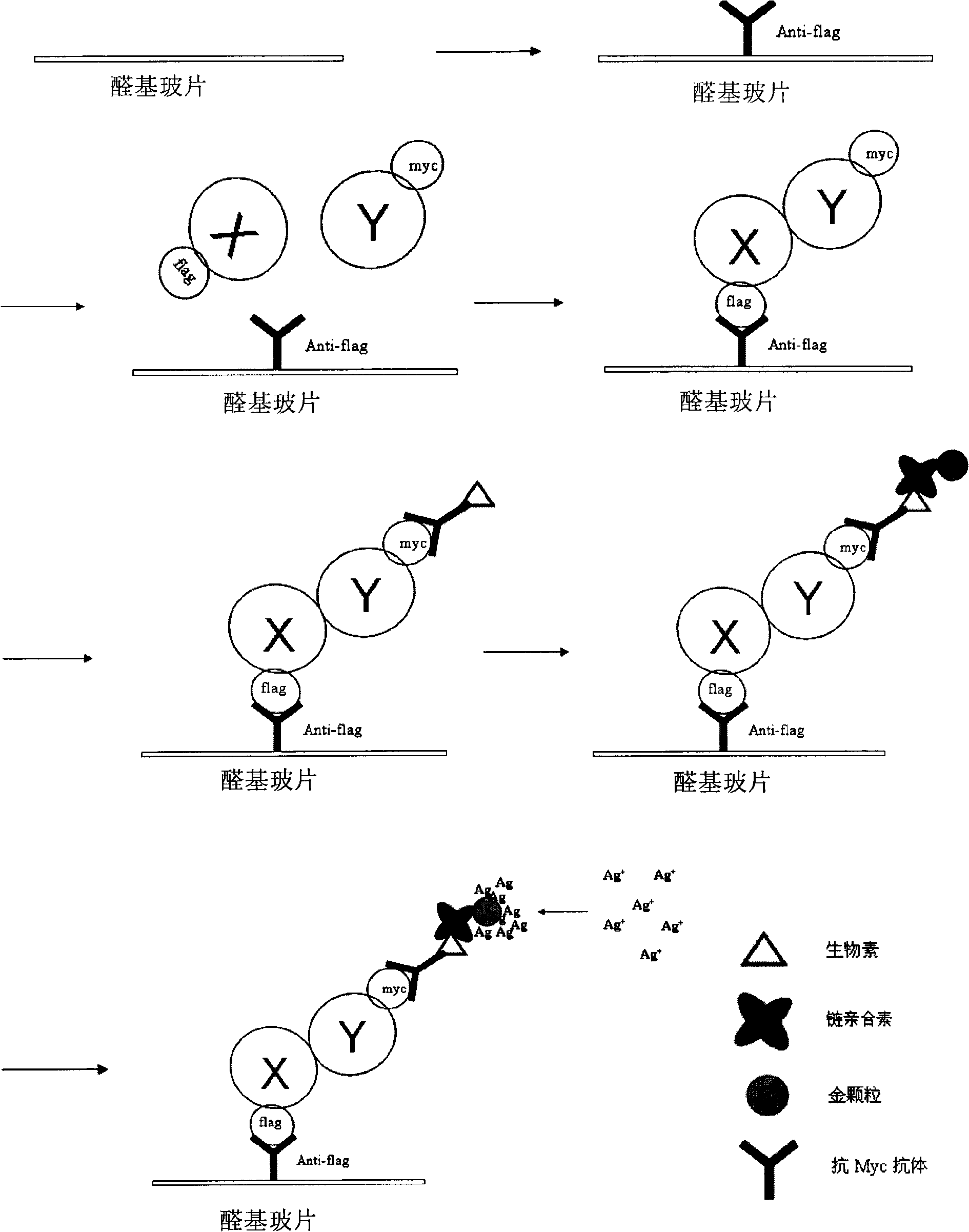
[0064] Preparation of the Flag antibody chip: Paste a film on the aldehyde-based glass slide to form 3 × 6 chip frames for differentiating samples. In each chip frame, spray anti-flag M2 monoclonal antibody and mouse IgG (as a control antibody) onto the aldehyde-based glass partition frame respectively, and dot three anti-flag M2 monoclonal antibodies in parallel in each reaction frame. Antibody and corresponding number of spots containing mouse IgG (forming a 2×3 array). Store at 4°C for later use.
[0065] In the embodiment of the present invention, the M2 monoclonal antibody was used. Since the M2 monoclonal antibody is a mouse IgG, mouse IgG was used as the antibody control. The present invention is not limited to the use of M2 monoclonal antibody, and other types of monoclonal antibodies can also be selected.
[0066] Construction of bait protein and prey protein expression vectors: the coding gene of bait protein was constructed on the flag tag vector, and the coding g...
Embodiment 1
[0069] Example 1: Co-immunoprecipitation chip analysis of the interaction between p65 and p50 subunits of nuclear transcription factor-κB (NF-κB)
[0070] The p65 and p50 subunits of NF-κB exist in the form of heterodimers under physiological conditions. Therefore, this example is designed based on the interaction between p65 and p50 as a positive model.
[0071] 1. Preparation of the Flag antibody chip: Paste a film on the aldehyde-based glass slide to form 3×6 chip frames in partitions. Use a microarray spotter (microarrayer, CAPITALBIO Boao, Jingxin SmartArrayer-48) to spray anti-flag M2 monoclonal antibody and mouse IgG (as a control antibody) onto the aldehyde-based glass slide frame in each chip frame In each reaction frame, 3 dots containing anti-flag M2 monoclonal antibody (upper row) and 3 dots containing mouse IgG (lower row) formed a 2×3 dot matrix. Prepare at least two Flag antibody chips and store them at 4°C for future use.
[0072] Two, the construction and id...
Embodiment 2
[0118] Example 2. Using the high-throughput visualization chip detection technology of the present invention to verify the interaction of 19 pairs of proteins with potential interactions, and compare the detection effect with the detection method of fluorescently labeled antibodies
[0119] In order to check the reliability of the high-throughput visualized silver-enhanced chip detection method of the present invention, 19 pairs of proteins with potential interactions (see Table 1) were randomly selected to verify with the high-throughput visualized chip detection technology of the present invention, and simultaneously adopted Fluorescence immunoassay method was tested, and each group of experiments carried a known positive interaction pair Flag-p50 / c-Myc-p65 and a known negative interaction pair Flag-Jun / c-Myc-lacZ as positive controls and negative and compare the results of the two sets of experiments.
[0120] The bait was respectively constructed on the pFLAG-CMV-2 eukaryo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com