KASP-based soybean core SNP markers and application thereof
A soybean and labeling technology, which is applied in the direction of DNA/RNA fragments, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of different seeds, high chip labeling density, high detection cost, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 Screening of soybean core SNP markers
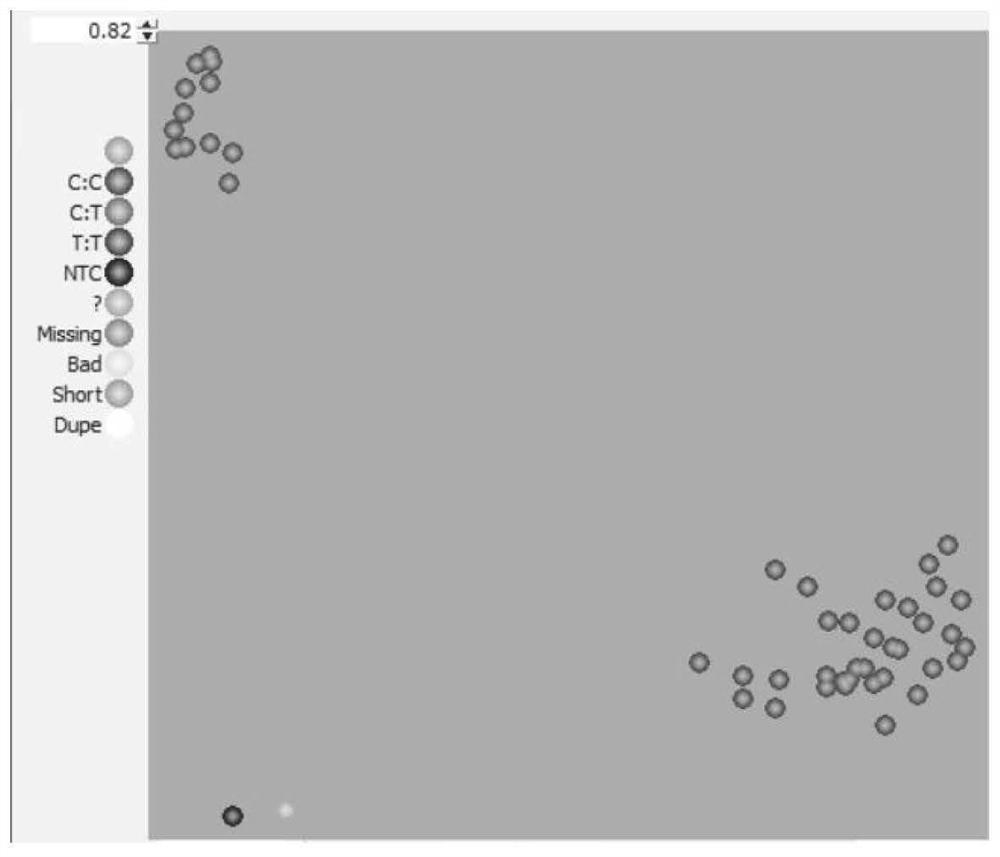
[0025] Through the detection results of 5,000 soybean resources by the "Illumina Zhongdouxin No. 1" chip, according to the principle of minimum allelic variation (MAF) ≥ 0.4, select SNP sites on different soybean chromosomes to ensure that all materials have a similarity <95% 65 high-resolution SNP sites were screened (Table 1), and KASP detection markers were developed and verified with 280 soybean resources.
[0026] Table 1 65 core SNP markers of soybean resources
[0027]
[0028]
[0029] The above SNP physical locations were determined based on the reference genome Wm82.a2.v1 (Phytozome, https: / / phytozome.jgi.doe.gov / pz / portal.html).
Embodiment 2
[0030] Example 2 A method for identifying 280 soybean resources using 65 soybean core SNP markers
[0031] (1) DNA preparation
[0032] Genomic DNA was extracted from 280 soybean leaves or soybean flour by CTAB method.
[0033] (2) Design and synthesis of KASP primers
[0034] The primer working solution was prepared by mixing three primers (Table 2), and the concentration of each primer was: AlleleFAM primer concentration 12 μM, AlleleHEX primer concentration 12 μM and universal primer (Common) concentration 30 μM. The 5' end of the AlleleFAM primer is labeled with FAM, and the 5' end of the AlleleHEX primer is labeled with HEX.
[0035] Table 2 KASP primer sequence (5'-3')
[0036]
[0037]
[0038]
[0039]
[0040] (3) PCR amplification
[0041] PCR system: PCR amplification-related reagents were added to the Array Tape, and the reaction system was shown in Table 3.
[0042] Table 3 PCR amplification system based on Douglas Array tape platform
[0043] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com