16S rDNA amplicon library building method for gestational diabetes mellitus intestinal flora detection
A technology of intestinal flora and amplicon, which is applied in the field of 16SrDNA amplicon library construction for the detection of intestinal flora in gestational diabetes mellitus, can solve the problems of high reagent cost, high proportion of additional testing samples, and low gel recovery efficiency. Achieve the effect of reasonable technical repeatability, good uniformity and rapid detection results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
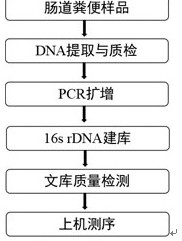
[0015] To achieve the above object, the present invention adopts the following technical solutions:.
[0016] Sample pretreatment
[0017] 1) Put about 300mg of feces sample into a 2ml centrifuge tube, add 1ml of PBS (0.1mol / L pH7.4), 20μL of 20%PVPP, shake and mix well
[0018] 2) Centrifuge at 2000r / min for 6min at room temperature, take the supernatant, add 1ml of PBS to the sediment, mix well, centrifuge, and take the supernatant;
[0019] 3) Combine the two supernatants, centrifuge at 12000r / min for 6min, and collect the precipitate.
[0020] Intestinal flora DNA extraction
[0021] 1) Add 300 μL Lysis Solution 1 (0.15mol / L NaCl 0.1mol / LNa 2 EDTA, pH8.0) 100mg / ml lysozyme 100μL 5μL proteinase K (10mg / ml), shaking at 500r / min for 30min at 37°C;
[0022] 2) Add 300 μL Lysis Solution 2 (10% SDS, 0.1mol / L NaCl, 0.5mol / L Tris-HCl, pH8.0), 50 μL 20% PVPP, keep at 65°C for 10 minutes, add 750 μL Tris-saturated phenol: chloroform: iso Amyl alcohol (25:24:1), mixed and shaken...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com