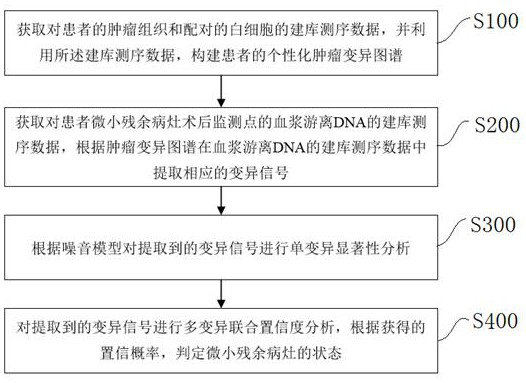
Tiny residual focus detection method and device, storage medium and equipment
A detection method and residual technology, applied in proteomics, medical data mining, instruments, etc., can solve the difficulties of judging the authenticity, unsatisfactory accuracy of small lesions, lack of correction methods, etc., to improve the credibility , Eliminate noise signals, and accurately detect the effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0145] Example 1: Performance analysis of hotspot-driven single mutation detection based on method 1
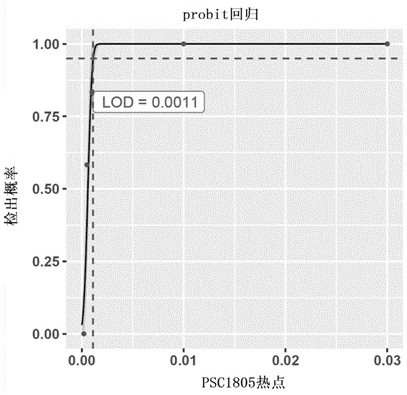
[0146]This example analyzes the experimental data for performance verification, based on the sensitivity and specificity of method 1 for hotspot-driven single variant detection. In this analysis performance verification experiment, the UMI (Unique Molecular identifier, UMI) molecular tag adapter was used to construct the library, and then the PanelP1 (Table 1.1) was used to enrich the target region. PanelP1 covered the 108Kb interval of 29 genes , the enriched library was subjected to high-depth sequencing. In the sensitivity assessment, 12 known hotspot-driven variations were used to make a standard positive sensitivity control-PSC1805 (see Table 1.2); in the specificity assessment, cfDNA from 149 healthy individuals was used to evaluate 19 Specificity of detection of driver variants in tumor hotspots.
[0147] 1.1 Sensitivity and minimum detection limit based on method 1 ...
Embodiment 2
[0169] Example 2: Performance Analysis of Single Variation Detection Based on Methods 1, 2, and 3
[0170] In this example, by analyzing the experimental data of performance verification, the sensitivity and specificity of the three analysis processes for the detection of non-hotspot single mutations are verified based on three different methods. The KAPA Hyper Preparation Kit (Roche Diagnostics product) was used to construct the library, and then the PanelP2 (see Table 2.1) was used to enrich the target region. PanelP2 covered the 2.1Mb interval of 769 genes. The enriched library was highly deep sequencing. During the performance evaluation, the samples used were prepared by mixing the white blood cell DNA of a single individual S with known SNP site information and the negative control negative standard GM12878.
[0171] 2.1 Sample information
[0172] The 32 SNP variations (single nucleotide mutations) of individual S different from hg19 (human genome version) and GM12878...
Embodiment 3
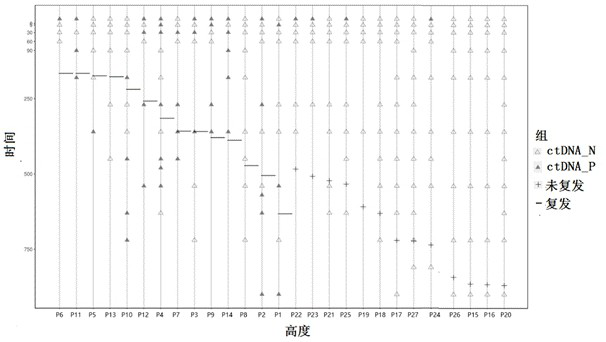
[0191] Example 3: Analysis of sample detection performance during multivariate tracking (based on method 1)
[0192] Since the content of cfDNA in blood limits the detection sensitivity of single mutations, method 1 can significantly improve the overall detection sensitivity by simultaneously tracking prior tumor-specific mutations in multiple tissues. In the MAVC2006 series of samples, different ratios of mixed DNA were used to simulate plasma DNA with different tumor proportions. In order to reduce the influence of site sampling, 100 random samplings were carried out by computer for each specified number, that is, 100 independent tumor prior variation maps were formed. For a diluted sample, each time according to each group The map carries out the mutation signal tracking of the designated site and determines the status of the small residual lesions. A total of 100 determinations are required. Finally, the positive detection rate of the 100 samples was counted as the detect...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com