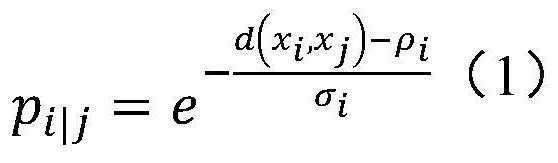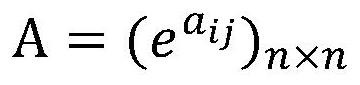Clustering method and system for unnormalized single-cell transcriptome sequencing data
A transcriptome sequencing, non-standardized technology, applied in the field of clustering methods and systems for non-standardized single-cell transcriptome sequencing data, can solve problems such as inaccurate clustering results and poor clustering results, and improve accuracy , the effect of improving performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
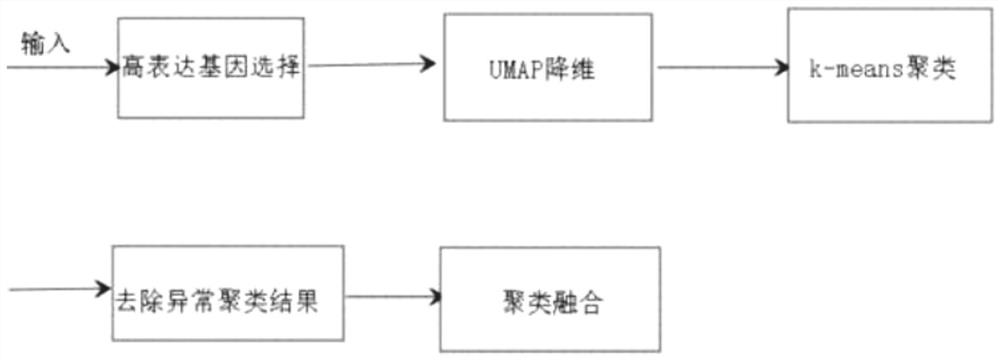
[0041] like figure 1 As shown, Embodiment 1 of the present disclosure provides a clustering method for non-standardized single-cell transcriptome sequencing data, including the following processes:
[0042] S1: Single-cell RNA sequencing data is stored in a matrix. The two dimensions of the matrix represent cells and genes, respectively, and the matrix value represents the expression of a gene in a cell.
[0043] After obtaining the input matrix, first select genes that are differentially expressed between cells for subsequent analysis, specifically, select some genes with higher coefficients of variation (mean divided by variance).
[0044] S1: Use UMAP to perform dimensionality reduction analysis on the matrix processed in S1.
[0045] In the high-dimensional part, the following formula is used to model the similarity between cells:
[0046]
[0047] where ρ i is the distance from the closest data point to i, and d can be any generalized distance that satisfies symmetr...
Embodiment 2
[0069] Embodiment 2 of the present disclosure provides a clustering system for unstandardized single-cell transcriptome sequencing data, including:
[0070] a data acquisition module, configured to: acquire single-cell transcriptome sequencing data;
[0071] a preprocessing module, configured to: preprocess the acquired sequencing data;
[0072] The preliminary clustering module is configured to: perform dimensionality reduction and clustering processing on the preprocessed sequencing data to obtain clustering results;
[0073] The clustering elimination module is configured to: arrange the clustering results from small to large or from large to small according to the Spearman correlation, and delete the clustering results with the small Spearman correlation from the gap where the Spearman correlation changes the most;
[0074] The hierarchical clustering module is configured to: take the average value of the equivalence relation matrix of each clustering result after deletio...
Embodiment 3
[0077] Embodiment 3 of the present disclosure provides a computer-readable storage medium on which a program is stored, and when the program is executed by a processor, implements the clustering of non-standardized single-cell transcriptome sequencing data oriented as described in Embodiment 1 of the present disclosure steps in the method.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com