Silkworm microsporidia milRNAs and application thereof
A technology for microsporidia and silkworm, which can be applied to DNA/RNA fragments, recombinant DNA technology, medical preparations containing active ingredients, etc. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1, Prediction and Analysis of Bombyx mori Microsporidia milRNAs
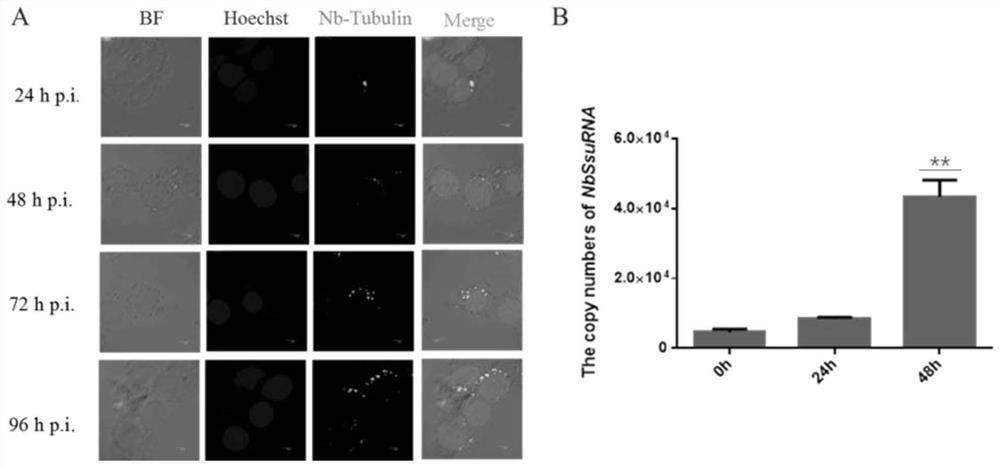
[0033]The proliferation characteristics of N. silkworm were analyzed by immunofluorescence and qPCR experiments, and the life history of N. silkworm was observed by immunofluorescence. It was found that when N. silkworm was infected for 24 hours, a small part of the spores had been ejected from the pole silk. The sporoplasm is injected into the host cytoplasm, and a small amount of sporoplasm can be found in the host cell; at 48 hours after infection, the sporoplasm of the spores becomes larger and develops into schizonts through binary or multiple divisions, which can be released in the host cell. The phenomenon of aggregation and expansion of sporoplasm was found; after 72 hours of infection, the mother spores formed by the fusion of schizonts into one formed sporocytes by binary division. At this time, it can also be observed in the host cells sporoplasm; by the time of infection for 96 hours, ...
Embodiment 2
[0083] Example 2, Identification and Functional Analysis of Bombyx mori Microsporidia milRNA-8
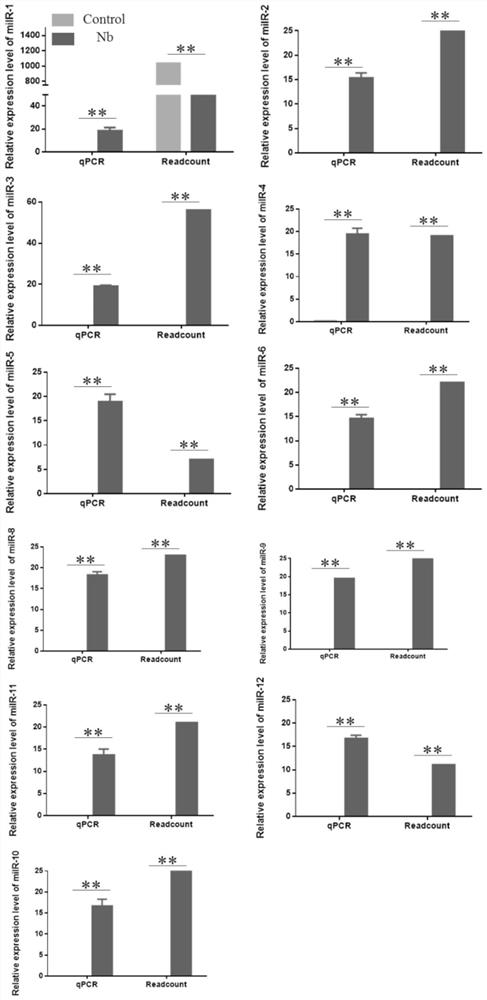
[0084] In order to explore the effect of milRNAs on the proliferation of Nos. silkworm, this example inserts target bands containing 10 milRNAs into puro-OpIE2 prm - In the mCherry-PA vector, the recombinant plasmid puro-OpIE2 was obtained by using the restriction enzyme Asc1 prm -mCherry-U6-milRNA( Figure 4 ), the results of further sequencing showed that 7 overexpression vectors were successfully constructed, and all of them were found to be expressed normally by fluorescence microscope observation. The results are as follows Figure 4 shown.
[0085] After the seven overexpression vectors were successfully constructed, they were transfected to BmE-SWU1 cells at the cellular level. After the overexpression vectors were stably expressed for 48 hours, N. silkworm was added. When the host cells were infected by N. silkworm for 48 hours, At this time, No. silkworm is in the stage...
Embodiment 3
[0088] Example 3, Screening and Identification of Bombyx mori Microsporidia milRNA-8 Target Gene
[0089] In order to identify the target gene of N. silkworm milRNA-8, this paper uses the target gene prediction software miRanda to make a preliminary prediction, and the miRanda threshold is set to score ≥ 140. The results showed that milRNA-8 predicted a total of 569 candidate target genes in the host, and selected 64 candidate target genes whose expression level difference between the Control group and the Nb group was ≥ 2, and performed KEGG functional enrichment analysis on these genes , 8 candidate target genes were found to be enriched in metabolism, neuroactive receptor-ligand pathway and peroxisome pathway. Corresponding quantitative primers were designed for 8 candidate target genes, and the sequences were: BGIBMGA002029-F: 5'-aagtatactgaaggccgagatg-3' (SEQ ID No.37);
[0090] BGIBMGA002029-R: 5'-tccatcaacacgtctcataaca (SEQ ID No. 38);
[0091] BGIBMGA009927-F: 5'-ctc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com