Screening and application of group of sulfonamide antibiotic broad-spectrum specific nucleic acid aptamers
A nucleic acid aptamer and antibiotic technology, applied in the fields of food safety and antibiotic detection, can solve the problems of single species of sulfonamide antibiotics, difficulty in meeting the needs of sample detection, etc., and achieves short screening period, high affinity and specificity, and simple chemical synthesis. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
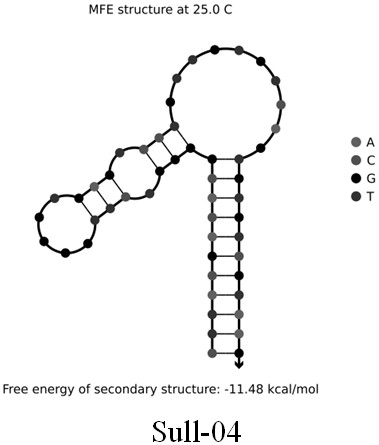
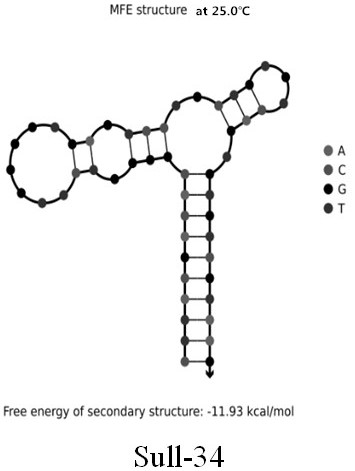
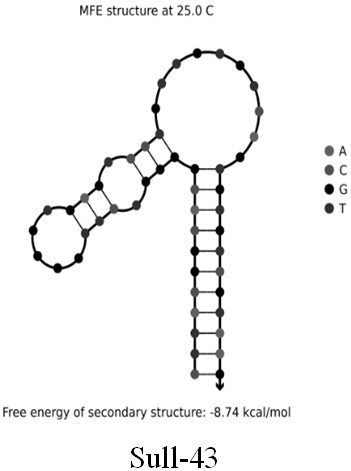
[0026] Example 1: Screening cloning, isolation, sequencing and prediction of secondary structure of single-stranded DNA for broad-spectrum specific nucleic acid aptamers of sulfonamide antibiotics
[0027] (1) Prepare 5 µL of a mixture of DNA library (5'-CGAGCATAGGCAGAACTTACGAC(N30)GTCGTAAGAGGCGAGTCATTC-3') and cDNA-biotin sequence (5'-TTTTTGTCGTAAGTTCTGCCATTTT / Biotin / -3') at a molar ratio of 1:5, and add to it Add 50 µL of 5X selection buffer (50 mM Tris-HCl, 2.5 mM MgCl 2 , 100mM NaCl, pH 7.4), and then use DNase / RNase free-water to make a 250µL mixture. After the mixture was placed in a metal bath at 95°C for 10 minutes, it was cooled at room temperature for more than 30 minutes to ensure the hybridization of the DNA library and cDNA-biotin sequences, thereby obtaining a DNA library-cDNA-biotin conjugate.
[0028] (2) Add 300 µL (200 µL at the beginning of the second round) of streptavidin-coated agarose magnetic beads (beads) to a 0.5 mL polyethylene bio-microcolumn (Bio-...
Embodiment 2
[0039] Example 2: PAGE analysis of the affinity of 3 sulfonamide antibiotic broad-spectrum specific nucleic acid aptamers to 19 commonly used sulfonamide antibiotics and other types of antibiotics
[0040] (1) Mix 90pmol of the enriched DNA library of the 11th round of screening with 450pmol of the cDNA-biotin sequence (5'-TTTTTGTCGTAAGTTCTGCCATTTT / Biotin / -3') solution in equal volumes and mix them in 227µL of 1X selection buffer (10mM Tris-HCl, 0.5mM MgCl 2 , 20mM NaCl, 0.01wt% Tween 20, pH 7.4), incubated at 95°C for 10min, and then cooled at room temperature for more than 30min to ensure the formation of DNA-enriched library-cDNA-biotin conjugates. At the same time, add 250 µL of beads solution to a centrifuge tube, wash with 1.5 mL of 1X selection buffer for 3 times, and centrifuge to discard the cleaning solution. Add the DNA-enriched library-cDNA-biotin conjugate into a centrifuge tube, and incubate on a sample mixer at room temperature for 30 minutes to immobilize the ...
Embodiment 3
[0046] Example 3: Fluorescence analysis of the affinity of 3 sulfonamide antibiotic broad-spectrum specific nucleic acid aptamers to 19 commonly used sulfonamide antibiotics and other types of antibiotics
[0047] (1) Contain 30µL of 20µM quenching probe BHQ-cDNA sequence (5'-GTCGTAAGTTCTG / BHQ1-3') and final concentration of 0.4µM aptamer Sull-04, Sull-34, Sull-43 sequence Add 30 µL of 0, 2, 4, 10, 20, 40, 100, 200, 400, 1000, 2000, 19 common sulfonamide antibiotics and 6 other types of the same type as in Example 2 of 30 µL antibiotic.
[0048] (2) Incubate the mixture obtained in step (1) at 72°C for 5 minutes, then incubate at 41°C for 5 minutes, then incubate at 25°C for 45 minutes, then take out 40 µL of the mixture and put it on the enzyme label plate Test its fluorescence spectrum on the instrument (excitation wavelength 480nm), and determine the effect of nucleic acid aptamers Sull-04, Sull-34, and Sull-43 on various antibiotics according to the fluorescence intensity...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com