Application of SEA in preparation of drugs for treating inflammatory bowel diseases and biomarker for detecting curative effect of inflammatory bowel diseases
A technology of inflammatory bowel disease and biomarkers, which is applied in the field of biomarkers to detect the efficacy of inflammatory bowel disease, and can solve the problems of slow curative effect, easy drug resistance, and increased treatment of inflammatory bowel disease
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
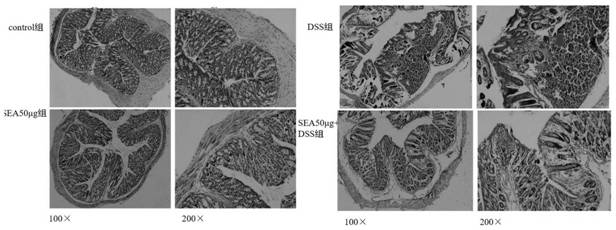
[0036] Construction of a mouse model of intestinal inflammation and administration methods (see figure 1 )
[0037] 1. Grouping and administration of mice:
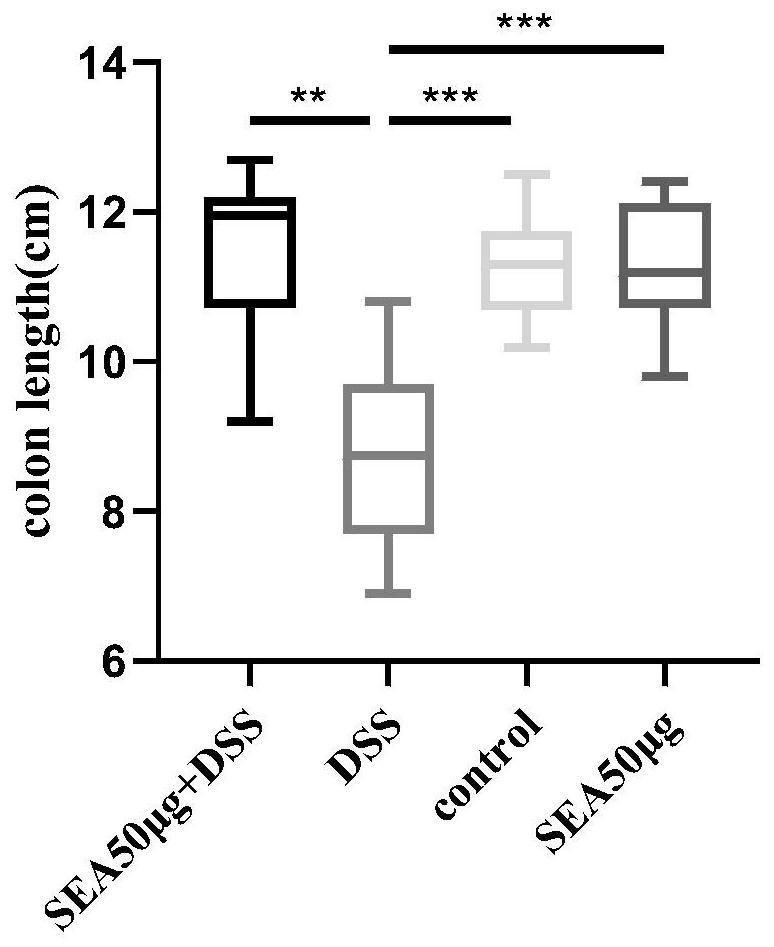
[0038] Forty mice (body weight 26.22±1.12g) were randomly divided into 4 groups, namely control group (10 mice), SEA50μg group (10 mice), DSS group (10 mice), DSS+SEA50μg group (10 mice).
[0039]On the 0th day of the experiment, the DSS group and the SEA50μg+DSS group were given 3% DSS (3g DSS per 100ml of drinking water) instead of normal drinking water. And on the 0th day of DSS modeling, each mouse in the SEA50μg group and SEA50μg+DSS group was given intraperitoneal injection of SEA50μg, while the control group had a normal diet.
[0040] The modeling method and the criteria for verifying the successful modeling can be found in the prior art: [1] BAUER C, DUEWELL P, MAYERC, et al. Colitis induced in mice with dextran sulfate sodium(DSS) is mediated by the NLRP3 inflammasome[J]. Gut , 2010, 59(9):1192-1199.
[0041...
Embodiment 2
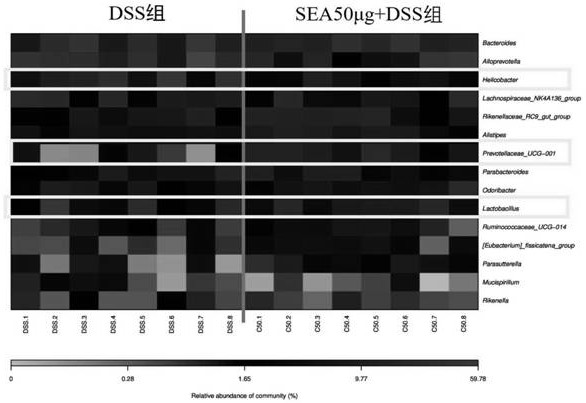
[0062] The feces of the mice on the 10th day of the experiment in Example 1 were used as materials, and the 16s rRNA universal primer was used to amplify and sequence the intestinal flora to explore the relationship between SEA, ulcerative colitis and intestinal flora. relation.
[0063] The bioinformatics analysis of the sequencing data is carried out as follows:
[0064] Raw data is in FASTQ format. Using Trimmomatic [1] The software de-scrambles the original paired-end sequence. The impurity removal parameters are: detect and truncate the ambiguous base N; and use the sliding window method to check the average base quality. When the quality is lower than 20, the previous high-quality sequence is intercepted. The double-ended sequence after decontamination utilizes FLASH [2] software. The splicing parameters are as follows: the smallest overlap is 10 bp, the largest overlap is 200 bp, and the largest mismatch rate is 20%.
[0065] To ensure the accuracy of the results,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com