COVID-19 and delta mutant strain detection kit and detection method thereof
A coronavirus and mutant technology, applied in the field of new coronavirus and its delta mutant detection kits, can solve the problems of prone to false negative results, poor accuracy, unfavorable prevention and treatment measures, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Embodiment 1 The preparation of each reagent
[0058] The purpose of the present invention is to screen the wild-type and delta mutant strains of the novel coronavirus, search for the full-length sequences of the ORF1ab and N genes of the novel coronavirus in GenBank, perform blast analysis on the ORF1ab and N genes separately, and compare them to obtain the target gene Conserved sequence segment, design the primer probe scheme in the conserved sequence segment of the gene, evaluate the Tm value, the difference between the Tm value of the target corresponding to the probe, GC content, avoid hairpin structure and dimer, etc. during the design process . It is used to distinguish the S gene of wild-type and mutant strains, search for the full-length sequence of the new coronavirus S gene in GenBank, perform blast analysis, select the L425R and T478K mutation sites, and try to avoid hairpin structures, primer internal dimers, The formation of dimers and mismatches between ...
Embodiment 2
[0066] Embodiment 2 Primer probe single verification
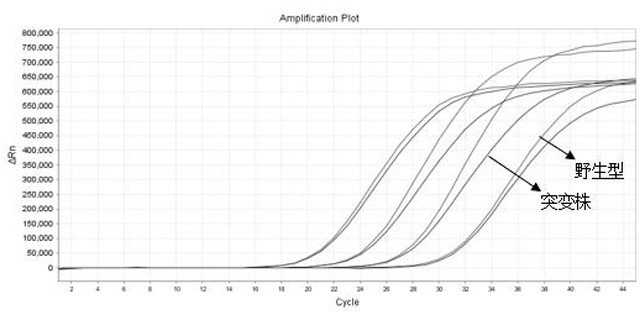
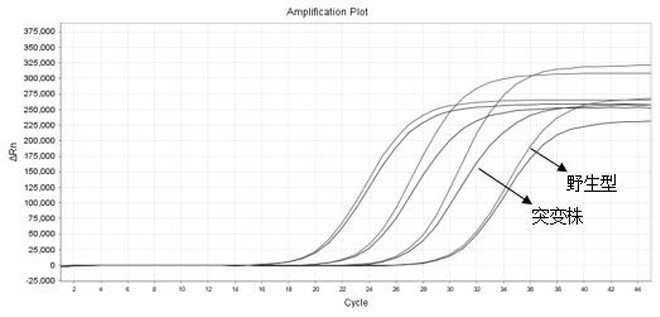
[0067] Take the wild-type pseudovirus, delta mutant pseudovirus and RP pseudovirus respectively, and use a commercial viral nucleic acid extraction kit to extract the nucleic acid to be tested as a template (4 concentration groups are set for each sample, corresponding to each target gene) PCR reaction system was prepared as follows: 15 μL of PCR reaction solution, 4 μL of primer-probe reaction solution (final concentration of upstream and downstream primers 50-500 nM, final concentration of probe 25-250 nM), Enzyme mixture 1 μL, template 5 μL, total volume 25 μL. Put the PCR tube into the fluorescence quantitative PCR instrument, and carry out the PCR reaction according to the following procedure: 50°C for 5 minutes; 95°C for 30s; 95°C for 5s, 66°C for 34s (collecting fluorescence), and cycle 45 reactions. The result is as Figure 1~Figure 4 and shown in Table 1.
[0068] Table 1 Amplification results of primer-probe s...
Embodiment 3
[0072] Embodiment 3 Mix fourfold
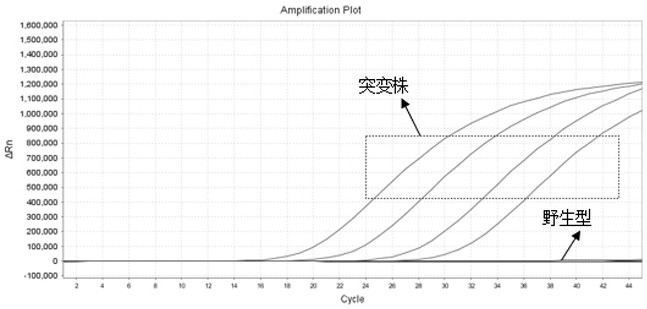
[0073] Four kinds of primer probes were combined into a quadruple system, and four different concentrations of delta mutant pseudoviruses were amplified, and the results were as follows: Figure 5 shown.
[0074] Table 2 Mixed quadruple validation amplification results
[0075]
[0076] *In order to simulate the real clinical sample collection process, according to past experience, the input concentration of fixed RP pseudovirus is 6×10 5 copies / mL.
[0077] This example illustrates that the quadruple system can detect different concentrations of delta mutant pseudoviruses, the amplification line is a standard S-shaped curve, and the Ct values of the 10-fold diluted pseudoviruses are evenly spaced.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com