Mixed sample detection method for detecting purity of Chinese cabbage seeds based on mSNP technology
A detection method, Chinese cabbage technology, applied in the direction of recombinant DNA technology, biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of cumbersome operation steps, high cost, complicated operation, etc., to improve detection efficiency, The effect of low cost and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
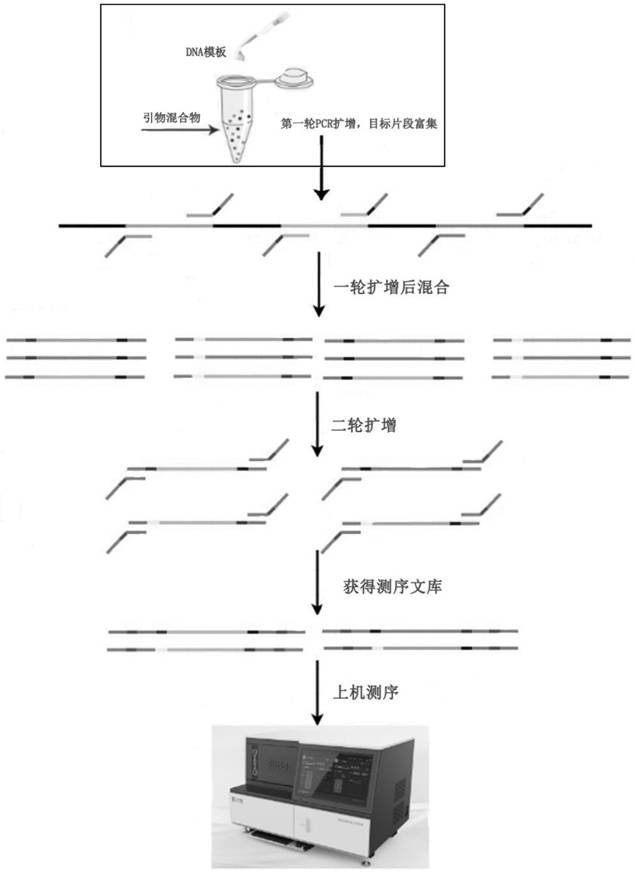
Image
Examples
Embodiment 1
[0089] Embodiment 1: Obtaining method of specific primer
[0090] The method for obtaining specific primers of the present invention is specifically:
[0091] Using the whole genome resequencing data of Chinese cabbage, use BWA-mem (http: / / bio-bwa.sourceforge.net / ) to post to the reference genome of Chinese cabbage, and use GATK (https: / / software.broadinstitute.org / gatk / ) for single nucleotide variant identification.
[0092] For the set of identified single nucleotide variation sites, screen the minimum allelic variation frequency > 0.02, heterozygosity rate < 15%, and deletion rate < 20%, combine the single nucleotide variation sites, and screen the single nucleotide The number of sites is located in the segment of 2-10, that is, the mSNP (polynucleotide polymorphism) site. Compared with the traditional SNP (single nucleotide polymorphism) site, the mSNP site can maximize the Using the information obtained by each primer pair, that is, as many SNP sites as possible can be...
Embodiment 2
[0095] Embodiment 2: Chinese cabbage seed purity detection primer set used
[0096] The primer set used for the detection of the purity of Chinese cabbage seeds includes the primer pair 1F / R~22F / R. In the primer pair 1F / R~22F / R, it not only includes the specific primer sequence shown in SEQIDNo.1~44, but also includes Universal primer sequence, the F-terminal universal primer sequence of the F primer in the primer pair 1F / R~22F / R is shown in SEQIDNo.45; the sequence of the R-terminal universal primer of the R primer in the primer pair 1F / R~22F / R As shown in SEQ ID No.46;
Embodiment 3
[0097] Embodiment 3: the obtaining method of primer mixture:
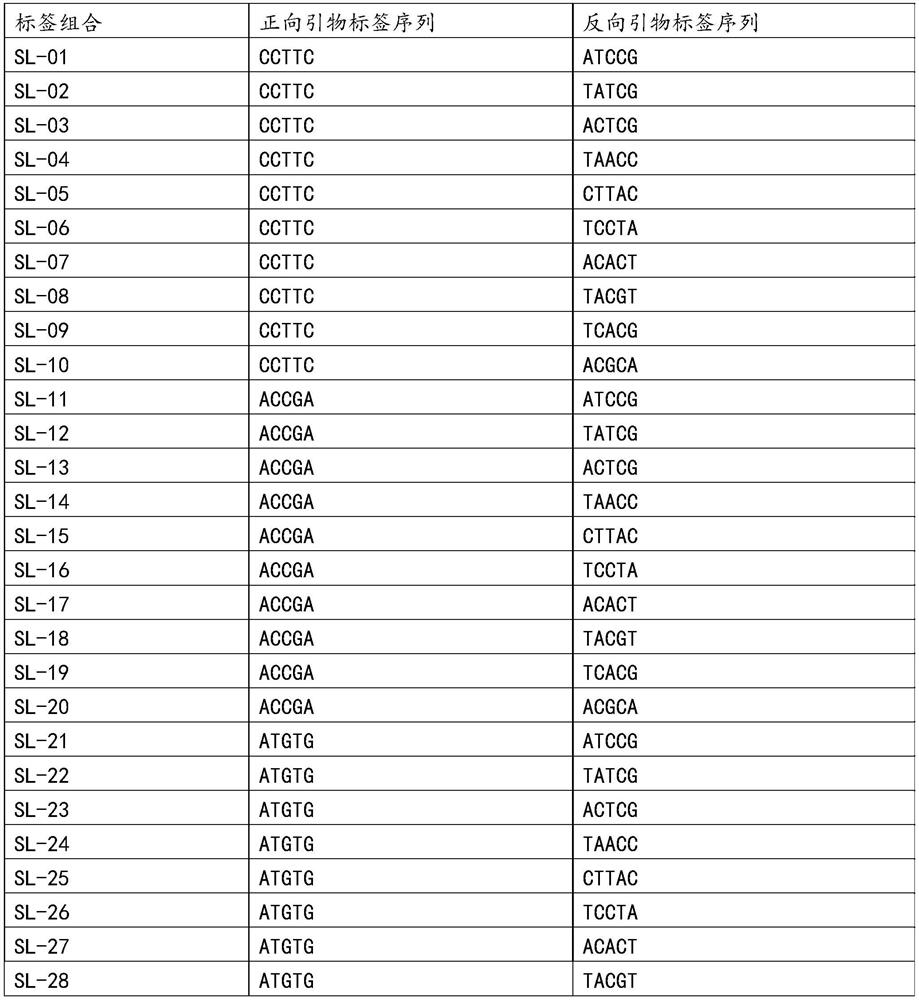
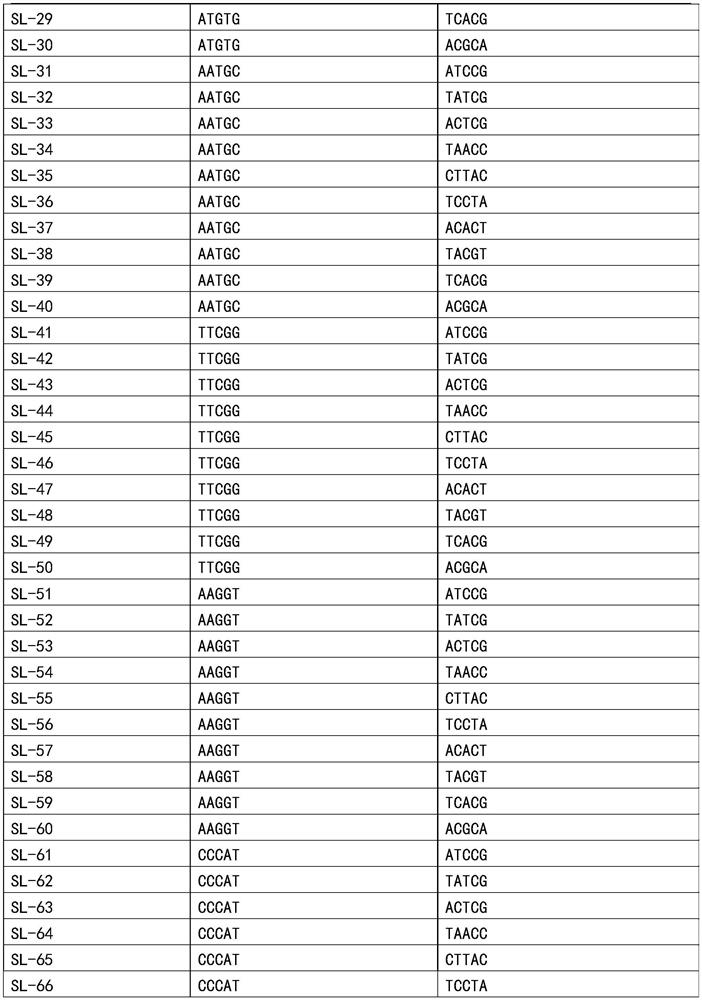
[0098] After obtaining specific primers, design specific tag sequences, and then re-synthesize primers. At this time, specific tag sequences will be added during primer synthesis. In this example, 96 sets of specific tag combinations are used, specifically:
[0099]According to the combination of specific tags, each specific primer was synthesized into 10 primers with different target tags. The sequence form of the primers with the target tags is shown in Table 2. And all R (reverse primers), take 10 μl of each primer, and dilute to 10ml; the concentration of each primer is 0.1 μM, and this embodiment prepares a total of 96 sets of primer pairs containing specific label combinations, that is, the primer mixture;
[0100] Table 2 Primer Set
[0101]
[0102]
[0103] "FF" is the F-terminal universal primer sequence, and the F-terminal universal primer sequence is AACGACATGGCTACGATCCGACTT, as shown in SEQ ID N...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com