Gene-deficient type Amycolatopsis sp. capable of highly producing vanillin, construction method and application thereof
A technique of Amycolatopsis and genetic defects, applied in the field of genetic engineering, can solve the problems of no research on secondary metabolism and less research on transcription factors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Example 1: Construction of ROK-TR1 gene deletion, gene complementation, and gene overexpression strains
[0065] On the NCBI website, the sequence of the transcription regulator ROK-TR1 of the ROK family in Amycolatopsis sp. ATCC 39116 strain was compared according to the gene sequence of Amycolatopsis sp.ATCC 39116. The gene sequence is shown in SEQ ID NO:1 , the encoded protein accession number is WP_020420567.1.
[0066] 1. Construction of Amycolatopsis bacterium with deletion of ROK-TR1 gene
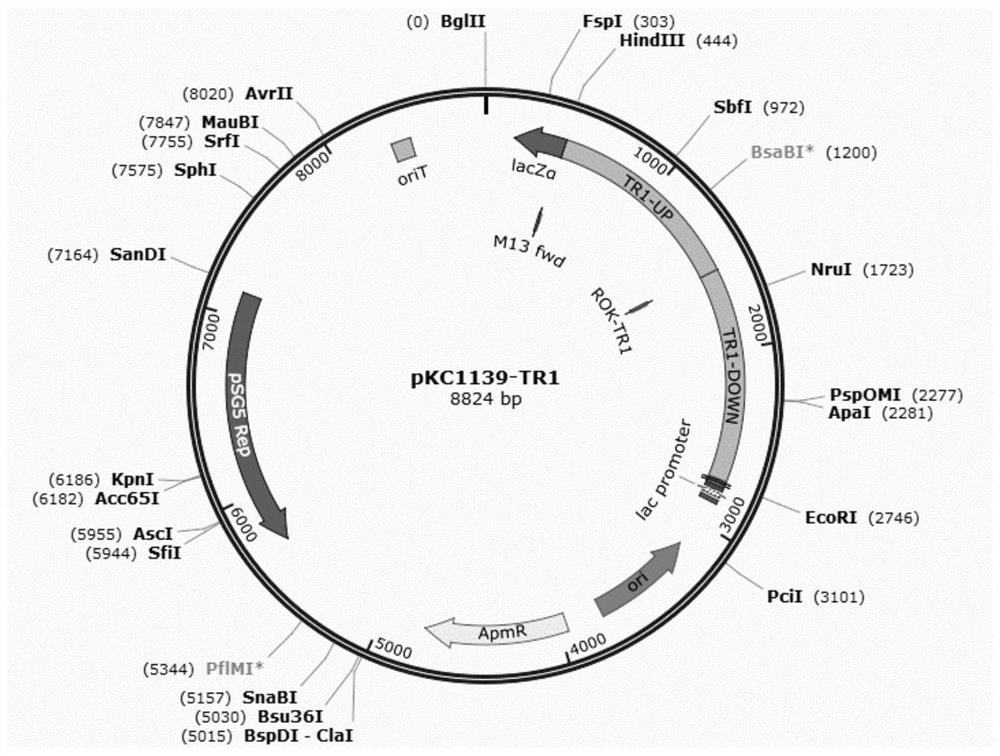
[0067] Using TR1-U-FOR / TR1-U-REV and TR1-D-FOR / TR1-D-REV as primers and Amycolatopsis genome as template, PCR amplifies the upstream and downstream of ROK-TR1 gene about 1.1kb each homology arm. At the same time, the above two TR1-UP and TR1-DOWN fragments were connected to the HindIII / EcoRI site of the pKC1139 plasmid through the Assembly kit to complete the construction of the plasmid pKC1139-TR1. The pKC1139-TR1 plasmid map is as follows figure 1 shown.
[0068] The pKC...
Embodiment 2
[0074] Example 2: Construction of ROK-TR2 gene deletion, gene complementation, and gene overexpression strains
[0075] On the NCBI website, the sequence of the transcription regulator ROK-TR2 of the ROK family in wild-type Amycolatopsis sp. ATCC 39116 was compared according to the gene sequence of the Amycolatopsis sp.ATCC 39116 strain. The encoded protein accession number is WP_020421841 .1.
[0076] 1. Construction of Amycolatopsis with ROK-TR2 gene deletion
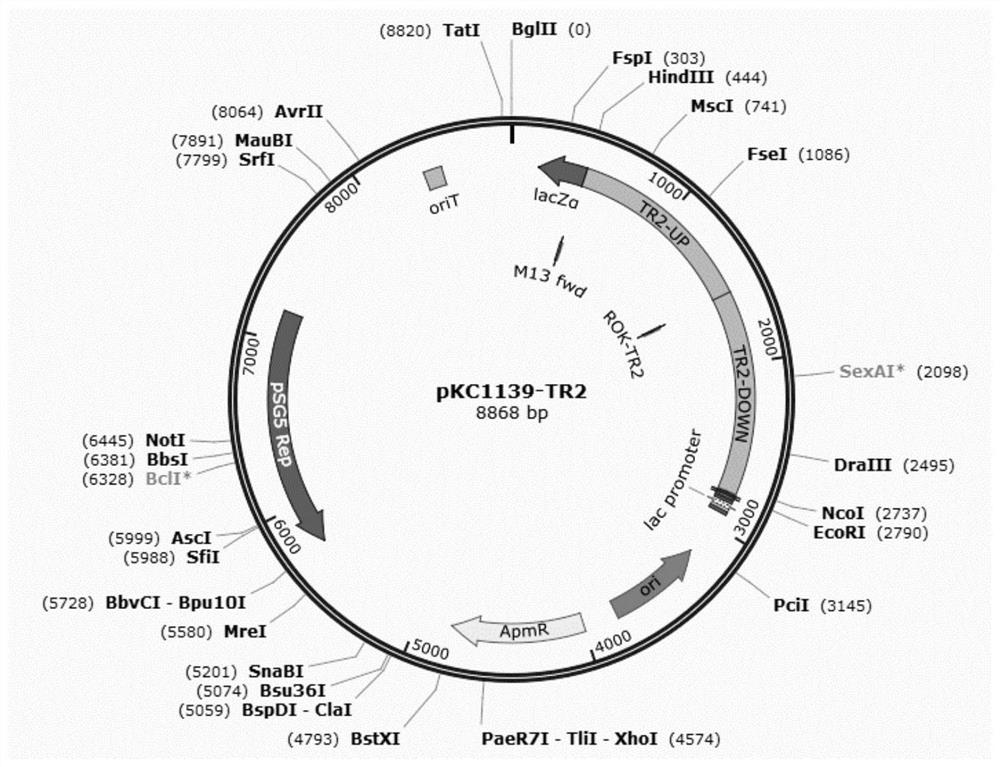
[0077] Using TR2-U-FOR / TR2-U-REV and TR2-D-FOR / TR2-D-REV as primers, Amycolatopsis genome as template, PCR amplifies the upstream and downstream of ROK-TR2 gene about 1.1kb each homology arm. At the same time, the above two TR2-UP and TR2-DOWN fragments were connected to the HindIII / EcoRI site of the pKC1139 plasmid through the Assembly kit to complete the construction of the plasmid pKC1139-TR2. The pKC1139-TR2 plasmid map is as follows figure 2 shown.
[0078] The pKC1139-TR2 plasmid was transformed into Amycol...
Embodiment 3
[0084] Example 3: Construction of ROK-TR3 gene deletion, gene complementation, and gene overexpression strains
[0085] On the NCBI website, the sequence of the transcription regulator ROK-TR3 of the ROK family in Amycolatopsis sp. ATCC 39116 strain was compared according to the gene sequence of Amycolatopsis sp. .
[0086] 1. Construction of Amycolatopsis bacterium with deletion of ROK-TR3 gene
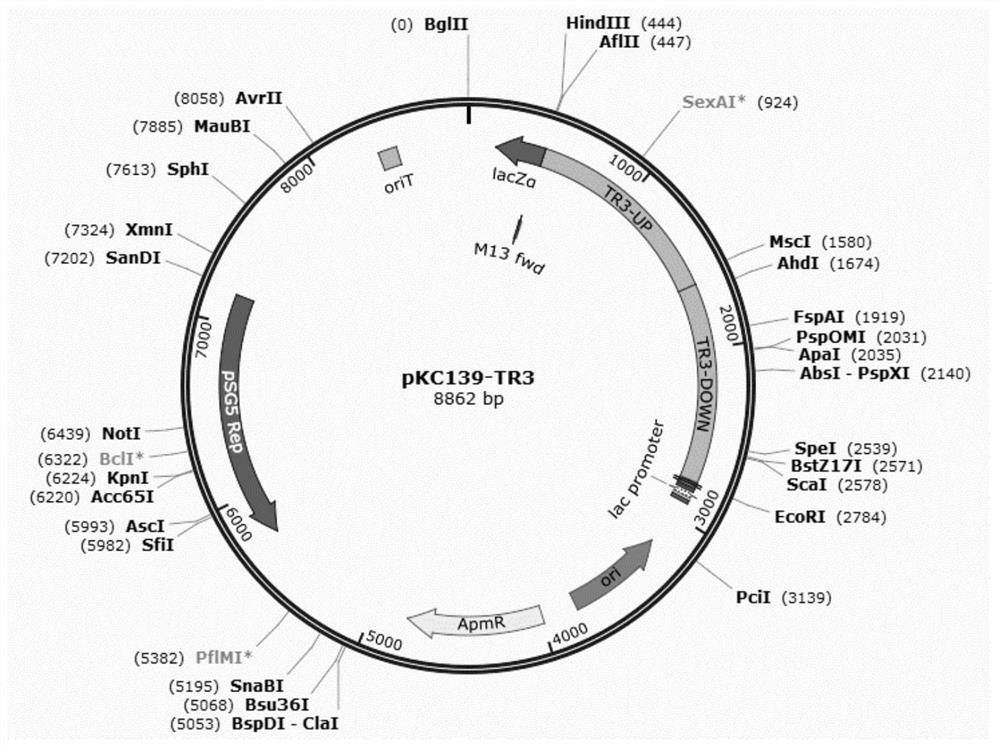
[0087] Using TR3-U-FOR / TR3-U-REV and TR3-D-FOR / TR3-D-REV as primers, and the Amycolatopsis genome as a template, PCR amplifies the upstream and downstream of ROK-TR3 gene about 1.1kb each homology arm. At the same time, the above two TR3-UP and TR3-DOWN fragments were connected to the HindIII / EcoRI site of the pKC1139 plasmid through the Assembly kit to complete the construction of the plasmid pKC1139-TR3. The pKC1139-TR3 plasmid map is as follows image 3 shown.
[0088] The pKC1139-TR3 plasmid was transformed into Amycolatopsis HM-141 by the method of conjugative transfer exper...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com