Primer group for identifying sika deer, red deer and hybrid deer, kit and application
A sika deer and primer set technology, applied in the biological field, can solve the problems of inability to identify hybrid deer antler, and achieve the effects of rich alleles, high polymorphism, and strong data sharing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0015] The embodiment of the present invention provides a primer set for identifying sika deer, red deer and hybrid deer. The primer set includes: upstream primers and downstream primers. Specifically, the primer set includes SEQ ID NO: 1 to SEQ ID in the sequence listing NO: 400 primers indicated.
[0016] Taking the publicly released red deer genome sequence GCA_002197005.1 as the reference genome, combined with the genome sequencing data of 100 sika deer and 68 red deer individuals obtained from the NCBI SRA database (acceptance number: PRJNA355630), using Samtools (Version 1.2) and BCFtools (Version:1.2) Identify the SNP sites on the genome, compare and analyze with the NT library of NCBI, and screen the MNP markers according to the following principles:
[0017] (1) The marker sequence is shared by sika deer and red deer, but not in other species;
[0018] (2) There are multiple discontinuous SNP differences in the sequence;
[0019] (3) The length of the marker sequenc...
Embodiment 2
[0039] The embodiment of the present invention provides an application of a MNP marker site, the application includes: the application includes: using the MNP marker site shown in SEQ ID NO: 401 to SEQ ID NO: 600 in the sequence listing for sika deer, horse Genetic difference analysis of deer and hybrid deer and pedigree analysis of sika deer, red deer and hybrid deer.
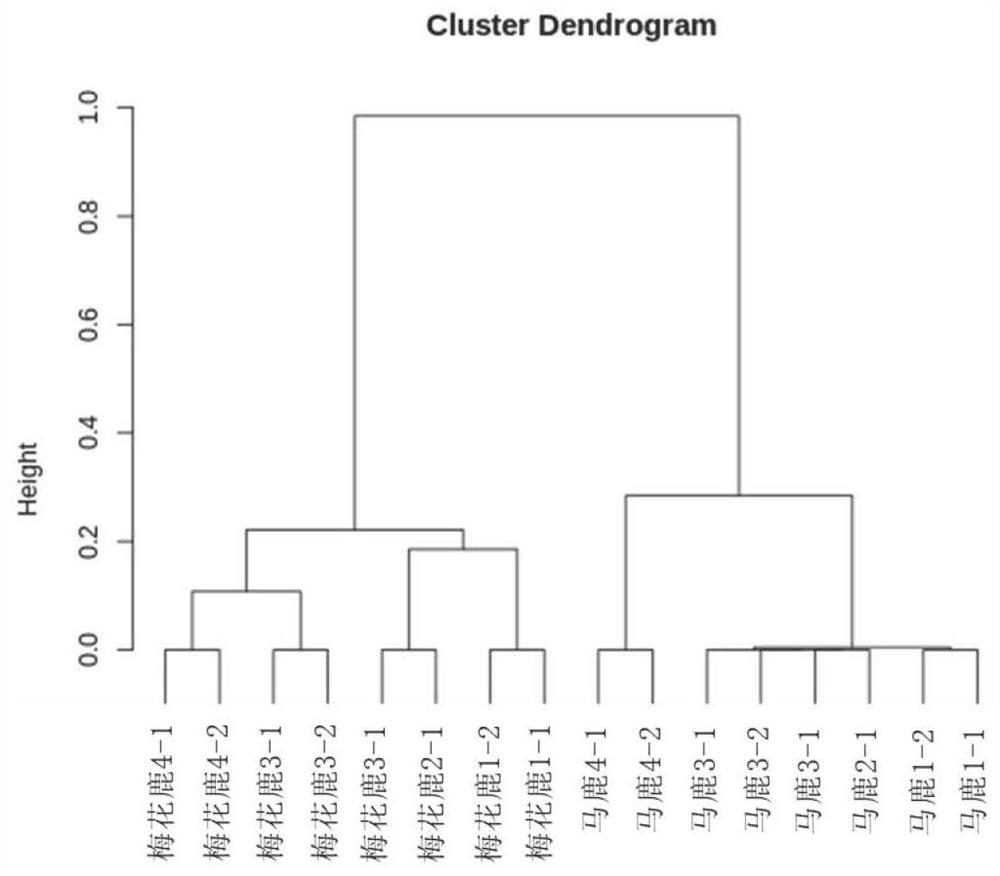
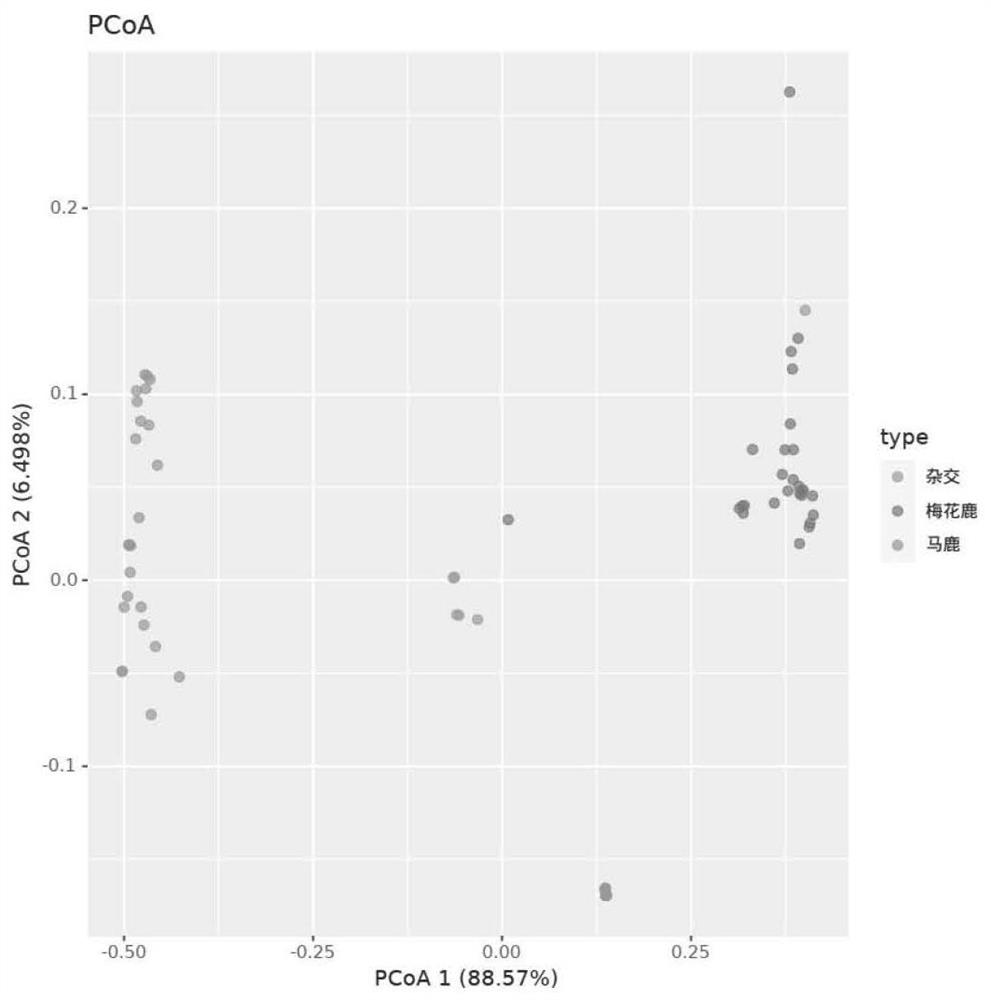
[0040] 29 sika deer, 32 red deer and 7 hybrid deer F1 generations donated by the Chinese Academy of Inspection and Quarantine using the 200 MNP marker sites and kits provided by the embodiment of the present invention, a total of 68 deer individual DNA were MNP marked Site difference analysis, through cluster analysis, it was found that all samples can be divided into three categories: red deer, sika deer and hybrid deer. figure 2 shown. Among them, 14 samples of red deer and 2 samples of sika deer and hybrid deer were grouped together, which is presumed to be caused by the wrong labeling of the sample sourc...
Embodiment 3
[0041] Example 3 Sika deer, red deer, and hybrid deer individual analysis of sika deer bloodlines
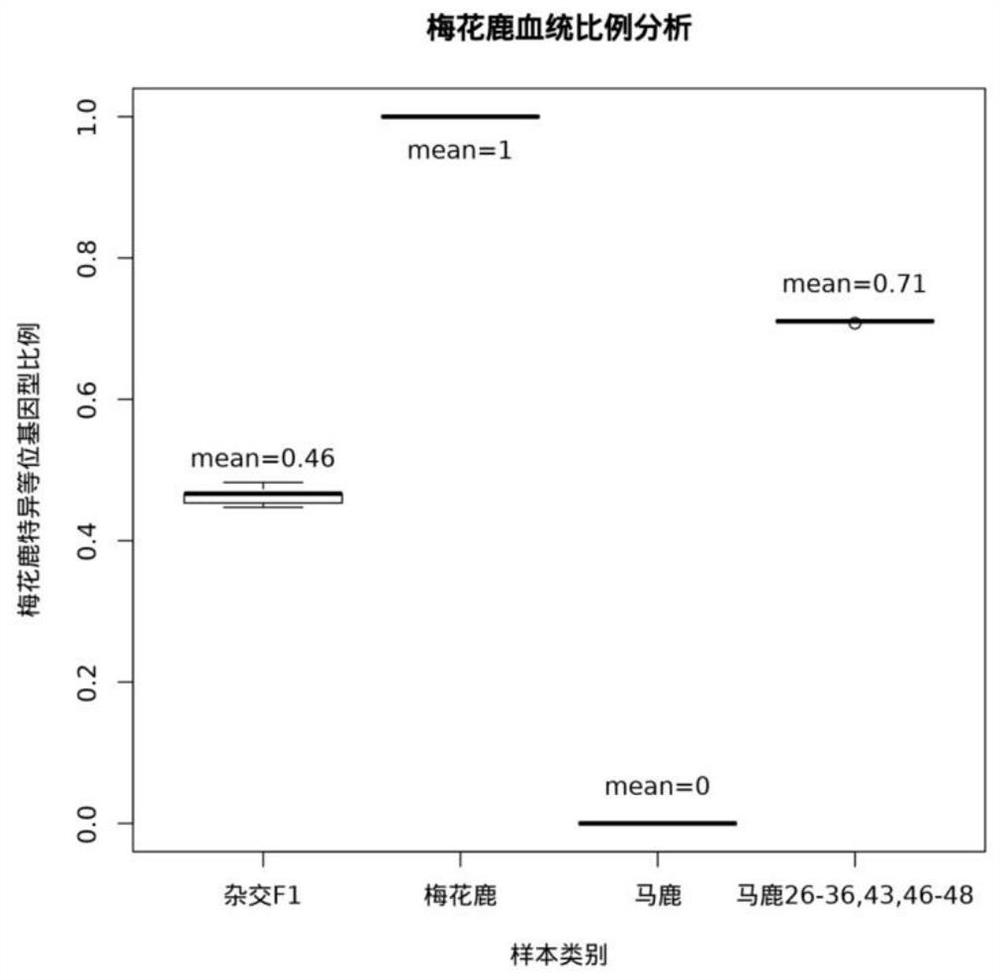
[0042] Excluding the sample sequencing data whose sample name was inconsistent with the classification in Example 2, the MNP marker typing data of the remaining 26 sika deer and 25 red deer were analyzed, and the specific MNP allele types that only existed in sika deer were screened. Obtain 78 MNP marker sites, analyze the number of sika deer specific allele types (T) at these 78 sites in the above samples, calculate the specific allele type ratio of sika deer in each sample, and establish the sika deer pedigree calculation method, namely Sika deer pedigree ratio (M) = number of sika deer specific alleles (T) / total number of 78 MNP marker alleles (N), the higher the M value, the closer the sika deer blood relationship. Utilize this method to carry out sika deer lineage analysis to 65 parts of deer individuals in embodiment 2, analysis result is as follows image 3 shown by im...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com