Digital PCR detection method for human PIK3CA gene mutation and application
A technology of use and reaction system, applied in the field of digital PCR detection of human PIK3CA gene mutation, can solve the problems of unsatisfactory sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
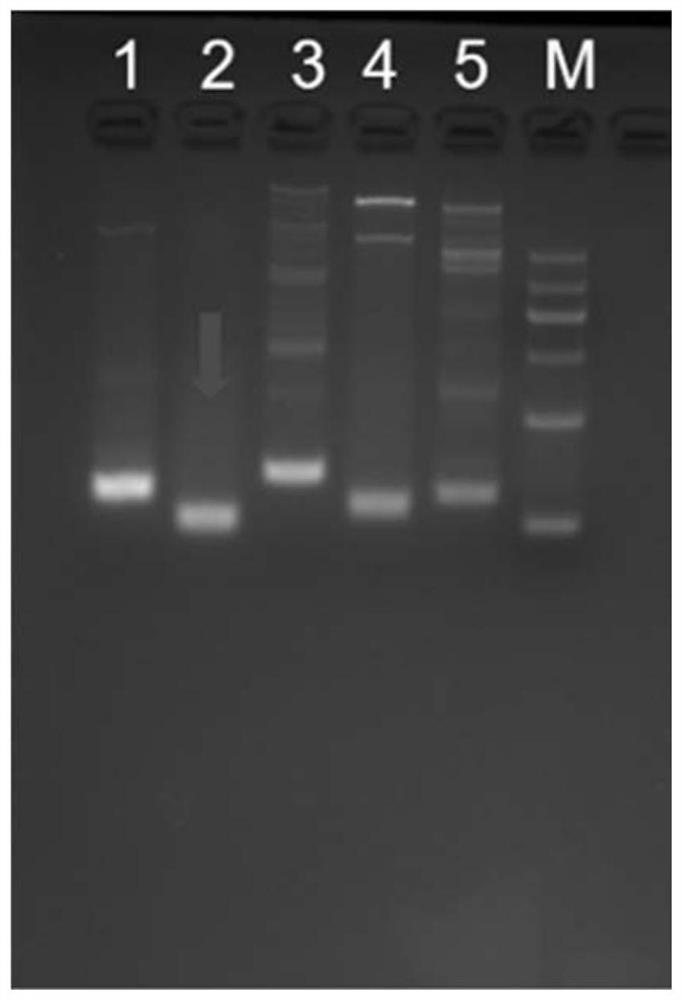
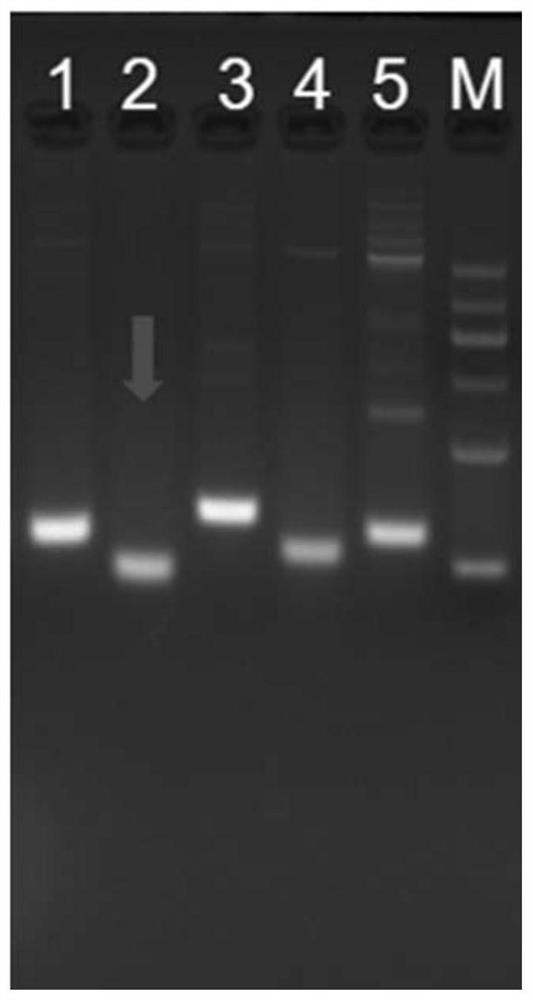
[0208] The screening of embodiment 1 primer pair
[0209] According to the sequence of the 9th exon of PIK3CA (E545, E542 sites), design 5 pairs of primers (primers were synthesized from Shanghai Sangon Biotechnology Co., Ltd.), the specific sequences are shown in Table 3.
[0210] Table 3. Screening 5 pairs of primers for amplification of PIK3CA E545, E542
[0211]
[0212]
[0213] According to the sequence of exon 20 of PIK3CA (H1047 site), 5 pairs of primers were designed (primers were synthesized from Shanghai Sangon Biotechnology Co., Ltd.), and the specific sequences are shown in Table 4.
[0214] Table 4. Screening of 5 pairs of primers used to amplify PIK3CA H1047
[0215]
[0216] Screening PCR system: 0.2ul of Taq HS polymerase, 2ul of 10×HS PCR buffer, 1.6ul of dNTP mixture, 1ul of upstream primer, 1ul of downstream primer, 5ul of tgDNA (human leukocyte genomic DNA, as a template), and make up the system with water to 20ul .
[0217] Screening PCR progr...
Embodiment 2
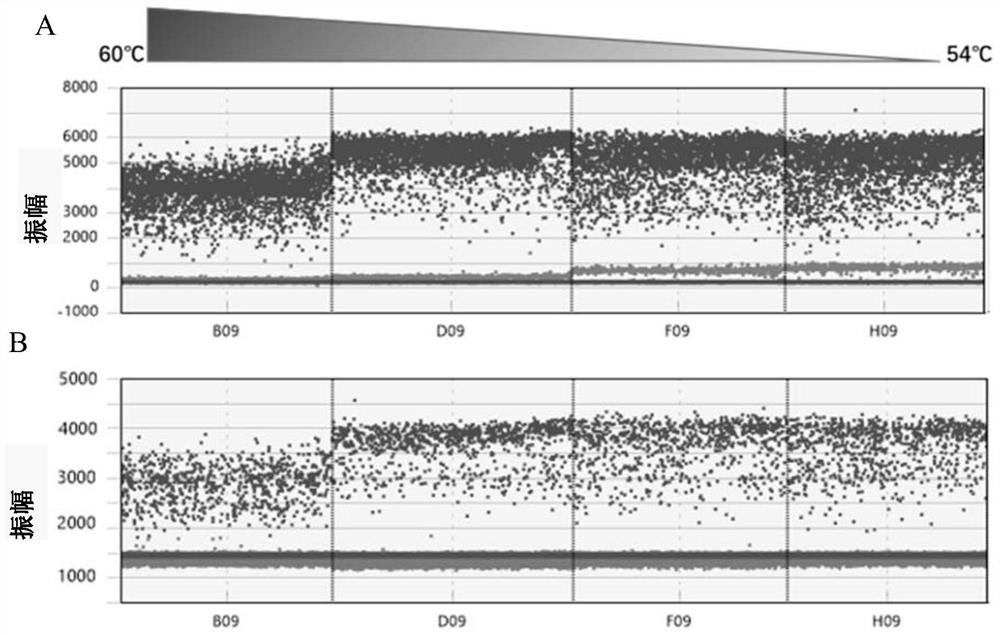
[0219] Embodiment 2 Taqman probe and the optimization of digital PCR reaction program
[0220] In this example, Taqman probes were synthesized and optimized, and the digital PCR reaction program was optimized based on the optimized Taqman probes.
[0221] 2.1 Taqman probe
[0222] In this embodiment, the detected PIK3CA gene mutations are nucleotide mutations corresponding to E545K, E542K, and H1047R (Table 5);
[0223] Table 5: PIK3CA gene mutation information table
[0224] Detection site mutation type COSMIC number genetic change E545K point mutation COSM763 c.1633G>A E542K point mutation COSM760 c.1624G>A H1047R point mutation COSM775 c.3140A>G
[0225] Based on the genome sequence and mutation site of human PIK3CA, the corresponding Taqman probes were designed and optimized. The optimized Taqman probes contained specific locked nucleotides (+N), respectively WTP-E545 / MTP-E545K, WTP -E542 / MTP-E542K, WTP-H1047 / MTP-H104...
Embodiment 3
[0237] Example 3 Sensitivity Verification of Detection of PIK3CA Gene Mutation by Digital PCR
[0238] Calculate wild-type template copy number (copies / ul), mutant template genome copy number (copies / ul) and mutation rate (%) according to Example 1, and dilute both to 4000 copies / ul with TE. The tgDNA was mixed into the genome template containing the mutation, so that the ratio of the mutation template was 0.06%, 0.13%, 0.25%, 0.5%, and 1%, respectively, and a gradient dilution template was prepared.
[0239] Validate digital PCR system: 2×ddPCR Supermix (No dUTP) 11ul, primer 1 (E545 / E542-F, H1047-F) 1.1ul, primer 2 (E545 / E542-R, H1047-R) 1.1ul, probe 1 (WTP-E545, E542 / WTP-H1047) 0.55ul, probe 2 (MTP-E545K, MTP-E545K, MTP-H1047R) 0.55ul, template 5.5ul, add water to make up to 22ul.
[0240] Add templates in the sample preparation area in the following order: blank control, negative control, and serially diluted template. The blank control is water, the negative control is ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com