Method for evaluating bacterial conjugational transfer efficiency based on quantitative PCR (Polymerase Chain Reaction) technology
A technology of conjugation transfer and technology, applied in the direction of microorganism-based methods, bacteria, recombinant DNA technology, etc., can solve the problems of weak sensitivity, poor repeatability, and low accuracy, and achieve strong sensitivity, high accuracy, and good repeatability Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
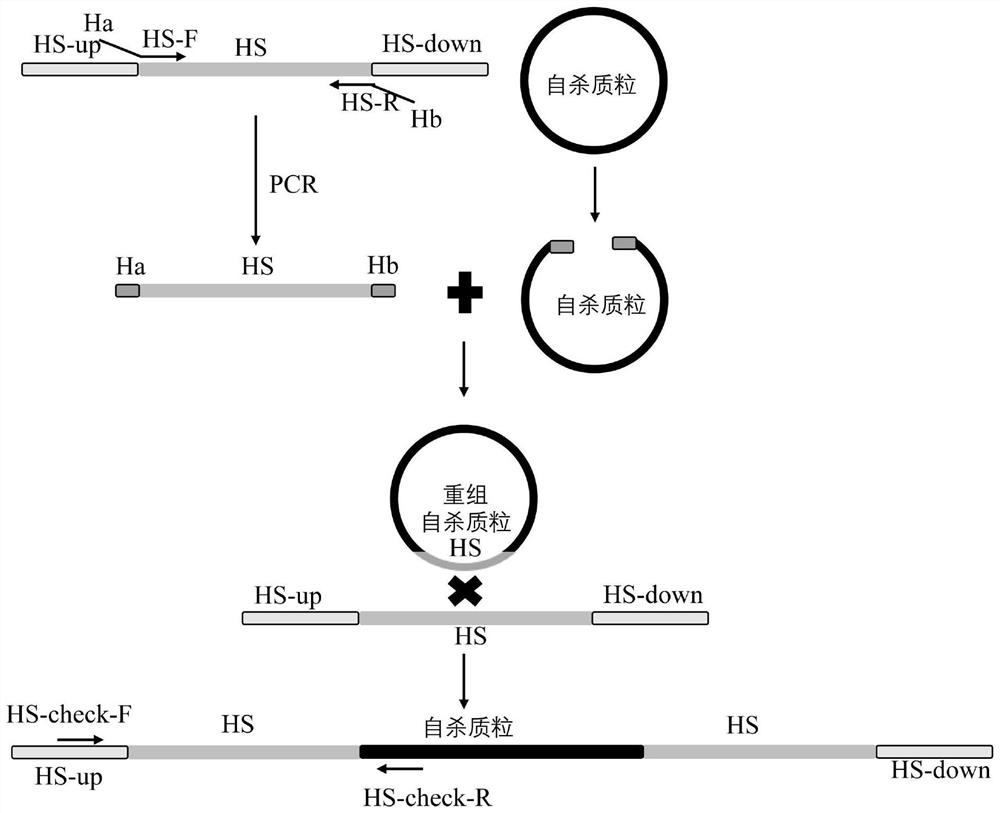
[0071] Taking the 1161031-1161687 and 1827872-1828471 homologous sequences on chromosome 1 in Vibrio harveylius 345 as an example to illustrate the construction of the bacterial conjugative transfer efficiency evaluation system by quantitative PCR ( figure 1 ).
[0072] The 1161031-1161687 sequence on chromosome 1 in Vibrio harveylius 345, with a length of 657bp, is located between the CU052_05900 gene and the CU052_05905 gene, belonging to the intergenic region; the 1827872-1828471 sequence on chromosome 1, with a length of 600bp, includes CU052_092703' end 128bp, CU052_09270 gene and the intergenic region of the CU052_09275 gene and the 151 bp of the 3' end of the CU052_092755.
[0073]In this study, the bacterial conjugative transfer efficiency evaluation system based on the quantitative PCR method of the homologous sequences of 1161031-1161687 and 1827872-1828471 on chromosome 1 in Vibrio harvelii 345 will be the key regulatory genes and influencing factors of horizontal g...
Embodiment 2
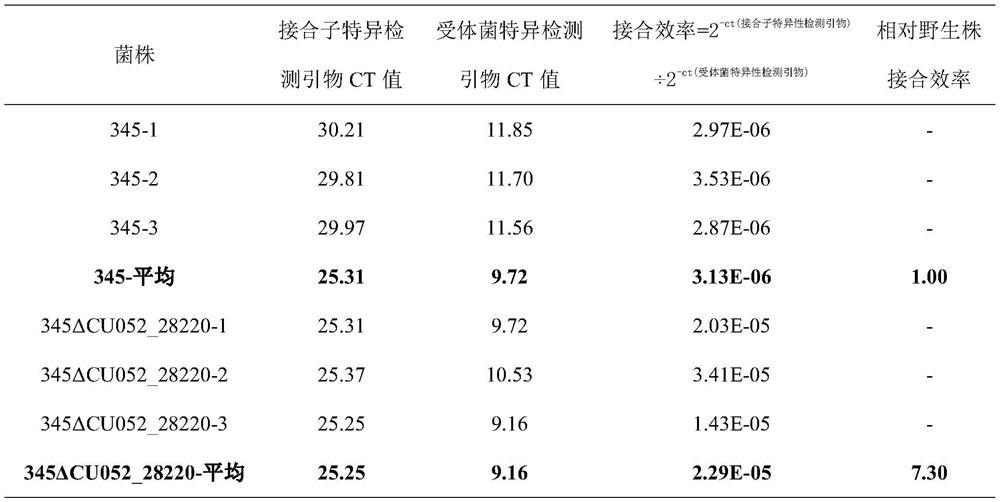
[0129] The bacterial conjugative transfer efficiency evaluation system based on the quantitative PCR method based on the homologous sequence of 1827872-1828471 on chromosome 1 in Vibrio harveylius 345 was used to analyze the logarithmic phase of Vibrio harveylius 345 and its gene knockout strain 345ΔCU052_28220 (GenBank number is AWB03109. 1) Conjugative transfer efficiency. Specific steps are as follows:
[0130] (1) Streak the donor bacterium pSW7848-HS2-E.coliGEB883 on an LB solid plate supplemented with 20 μg / mL chloramphenicol and 0.3 mMDAP (2,6-Diaminopimelic acid), and culture it overnight at 37°C for activation;
[0131] (2) Pick 3 monoclonal colonies formed by the activated bacteria in (1), inoculate them in LB liquid medium supplemented with 20 μg / mL chloramphenicol and 0.3 mM DAP, and cultivate to logarithm at 37°C and 200 rpm Early OD600nm=0.3~0.7;
[0132] (3) Configure an LBS solid plate, and activate the recipient bacteria Vibrio harvey 345 and 345ΔCU052_28220...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com