Method for detecting SARS-CoV-2 69-70del site based on RAA-CRISPR
A site-assisted detection technology, applied in the direction of microorganism-based methods, biochemical equipment and methods, DNA/RNA fragments, etc., can solve problems such as difficult to achieve rapid on-site detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Embodiment 1, be used for the design and preparation of crRNA of the present invention and PCR primer
[0063] 1. Design and preparation of crRNA used in the present invention
[0064] (1) Synthesis of primer sequences
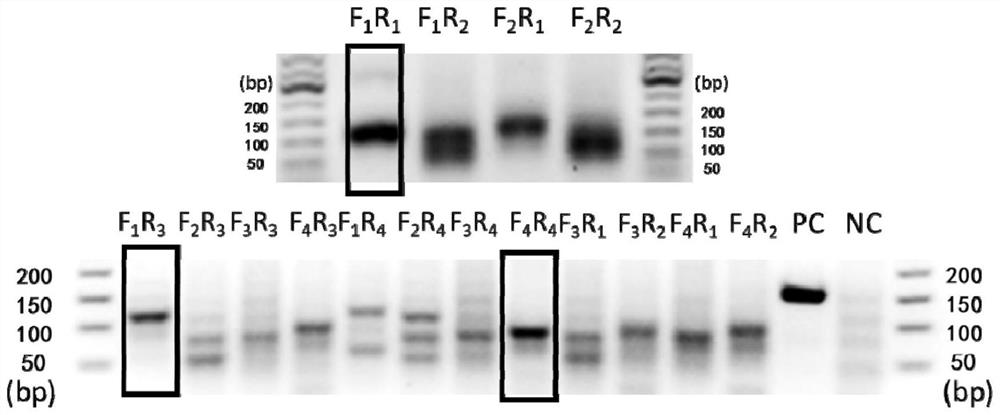
[0065] The present invention designs crRNA at the SARS-CoV-2 69-70del site and the wild site respectively. The 5' end of the crRNA has a 39nt repeat sequence, which can bind to LwCas13a protein, 5'-GGGGAUUUAGACUACCCCAAAAACGAAGGGGACUAAAAC-3', the single-stranded DNA sequence designed as a template is repeat sequence + target sequence, T7 sequence (5'- TAATACGACTCACTATAGGG-3') + partial repeat sequence (5'-GATTTAGACTACCCCAA-3') was used as the upstream primer, and the reverse complementary sequence about 20 bp downstream of the target sequence was used as the downstream primer (Table 1). Each sequence in Table 1 was synthesized.
[0066] Table 1. Template sequence and PCR amplification primer sequence for preparing crRNA used for mutation detection
...
Embodiment 2
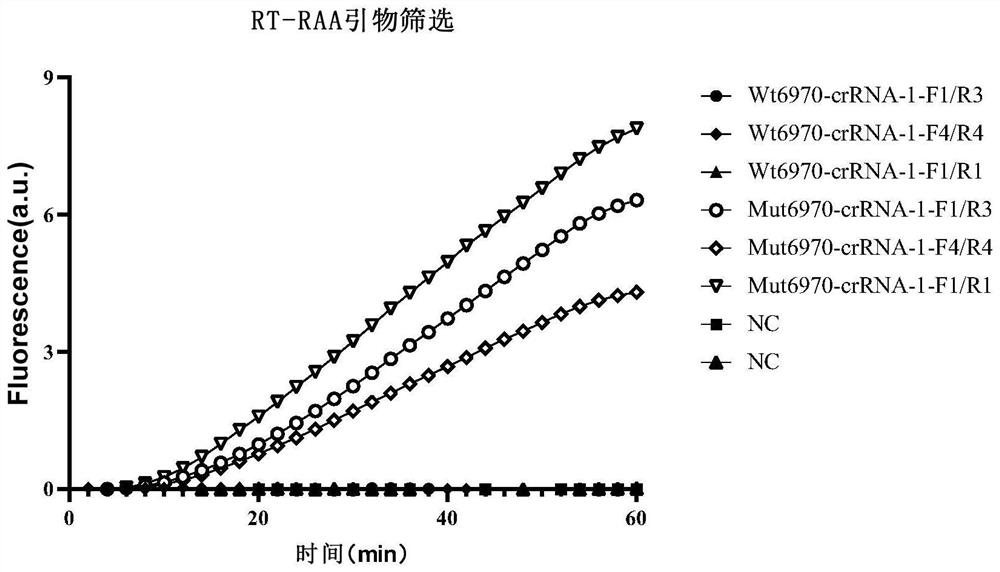
[0105] Example 2, Establishment of RT-RAA-CRISPR detection method for SARS-CoV-2 69-70del site
[0106] In this study, RT-RAA technology was used to amplify the target nucleic acid, and the target sequence detected by SARS-CoV-2 69-70del was transcribed to obtain the corresponding ssRNA, using the above crRNA: Mut6970-crRNA1, Mut6970-crRNA2, Mut6970 -crRNA3, Mut6970-crRNA4, Wt6970-crRNA1, Wt6970-crRNA2, Wt6970-crRNA3, Wt6970-crRNA4 were detected, the signal strength of different crRNAs was compared, and the crRNA with strong fluorescent signal and high sensitivity was selected as the subsequent crRNA detection. The specific steps and principles are as follows: the first step uses specific primers to amplify the target sequence (through denaturation, annealing, and extension processes), and there is a section of T7 transcription sequence at the 5' end of the primer, so that the PCR amplification obtained double Stranded DNA (dsDNA) is recognized and transcribed by T7 RNA polyme...
Embodiment 3
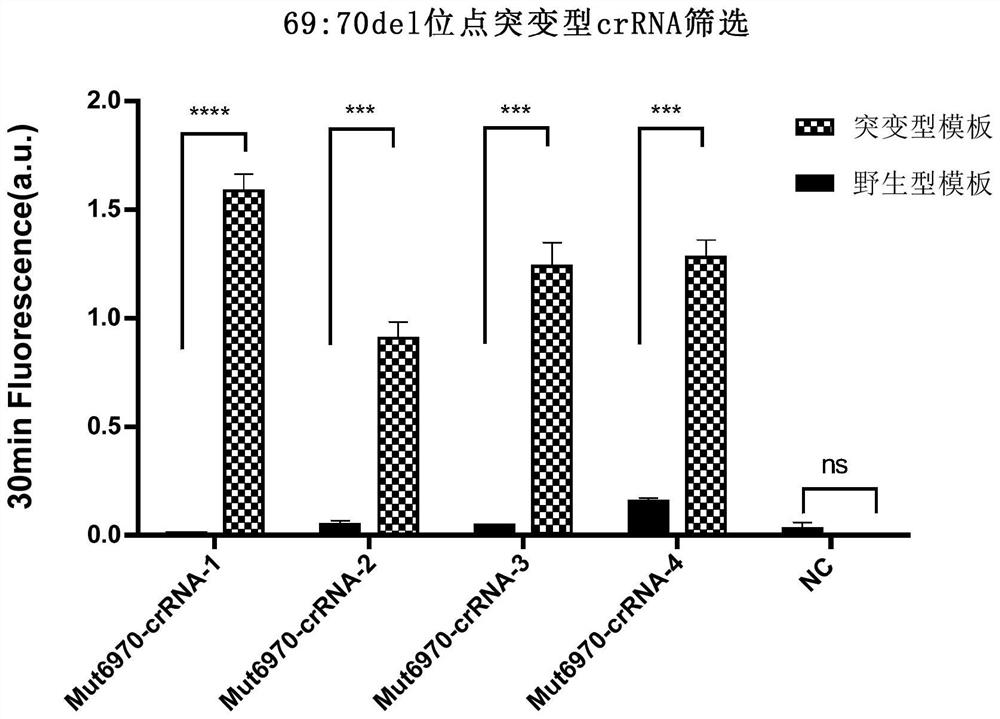
[0122] Example 3, Specificity and sensitivity detection of SARS-CoV-2 69-70del and wild crRNA
[0123] 1. SARS-CoV-2 69-70del optimal crRNA screening
[0124] In order to screen crRNA with higher detection sensitivity and shorter detection time, RT-RAA was performed using the wild-type and 69-70del mutant ssRNA constructed above and the designed primers 6970-RAA-F1 and 6970-RAA-R1 Amplify. And use 4 kinds of Mut6970-crRNA to detect the ssRNA of 2 sequences respectively.
[0125] (1) Synthesis of crRNA primer sequence
[0126] Each sequence in Table 1 of Example 1 was synthesized, and crRNA was synthesized and prepared according to the method of step 1 in Example 1.
[0127] (2) The mutant RNA template obtained in step 3 in Example 1 and the wild-type RNA template were respectively gradiently diluted to obtain RNA solutions containing different concentrations of 69-70del gene fragments and different concentrations of 69-70 wild gene fragments: 10 5 copies / μL, 10 4 copies / μ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com