Method for detecting SARS-CoV-2 69: 70del site
A sars-cov-269 and sars-cov-2 technology, applied in the field of molecular diagnosis, can solve the problems of time-consuming, high instrument precision requirements, and massive data analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Example 1, the design and preparation of crRNA and PCR primers for the present invention
[0068] 1. CRRNA design and preparation for the present invention
[0069] (1) Synthesis of primer sequence
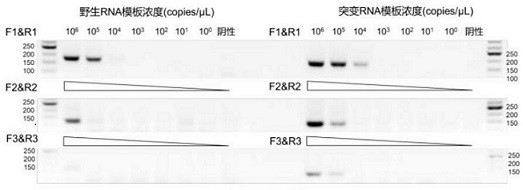
[0070] The invention is designed with wild sites in SARS-COV-2 69: 70DEL site. CrRNA's 5 'end with a repeating sequence of 39 nT, which can be combined with LWCas13a protein, 5'-gggauuuacuaccccaaaaaaaaaaaaagggguaaaaac-3', design as a single-stranded DNA sequence as a template to repeat sequence + target sequence, T7 sequence (5'- TaatacgActCACTATAGGG-3 ') + part repeat sequence (5'-GatttagActaccccaa-3') is an upstream primer, and the reverse complementary sequence of about 20 bp downstream of the target sequence is taken as a downstream primer (Table 1). The sequences in Table 1 are synthesized.
[0071]
[0072] (2) PCR amplification
[0073] The sequence shown in Table 1 is synthesized from Beijing Tianyi Huiyuan Company. According to the research method in the literature, ...
Embodiment 2
[0105] Example 2, PCR-CRISPR / Cas13a 69 detector 2 SARS-CoV-: establishing site method 70del
[0106] In this study, PCR amplification of the target nucleic acid, the SARS-CoV-2 69: 70del detected target sequence to be transcribed to give the corresponding ssRNA, with the above crRNA: Mut6970-crRNA1, Mut6970-crRNA2, Mut6970-crRNA3, Mut6970 -crRNA4, Wt6970-crRNA1, Wt6970-crRNA2, Wt6970-crRNA3, Wt6970-crRNA4 detected, comparing different signal strength crRNA choose a stronger fluorescence signal and a high sensitivity crRNA crRNA subsequent detection. Specific works as follows: The first step in using the target specific primer sequence (by denaturation, annealing, extension process), having a period of T7 transcripts of the primer sequences in the 5 'end, such that the double stranded DNA was amplified by PCR (dsDNA) can be transcribed and T7RNA polymerase recognition. The second step, the amplification product take-out portion of T7 RNA polymerase was added, LwCas13a protein can...
Embodiment 3
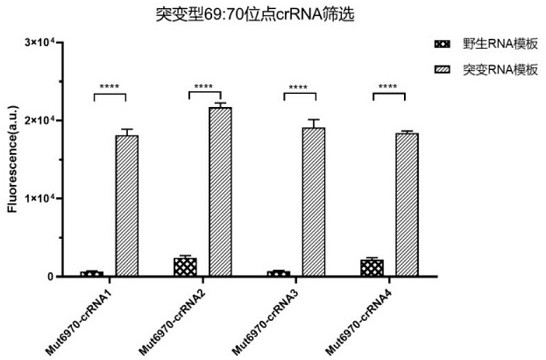
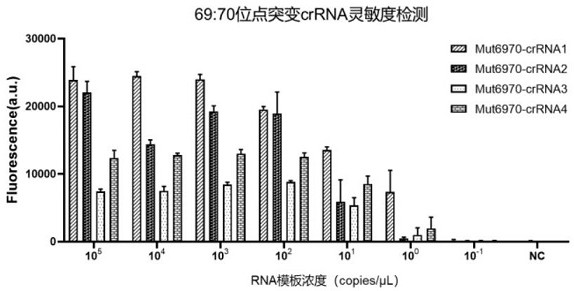
[0119] Example 3, SARS-CoV-2 69: 70del sensitivity and specificity of detecting wild crRNA
[0120] 1, SARS-CoV-2 69: 70del optimal screening crRNA
[0121] Was screened to a higher detection sensitivity, the detection time is shorter the crRNA, using the previously described construct wild-type, 69: 70del mutant ssRNA and design primers 6970-Primer 1-F, and 6970-Primer 1-R, PCR was performed amplification. And treated with 4 kinds Mut6970-crRNA ssRNA sequence detector 2, respectively.
[0122] Synthesis (1) crRNA primer sequence
[0123] Table 1 Synthesis of pieces of sequence, in Example 1, Step 1 of the synthesis method of Embodiment press and prepared crRNA.
[0124] (2) The procedure of Example 1 in the embodiment 3 mutant and wild-type RNA template RNA obtained were serially diluted to give 69 different concentrations: solution 70del RNA gene fragments with gene fragments of different concentrations of wild 69:70, followed by concentration 10 5 copies / μL, 10 4 copies / μL,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com