Specific primer pair for identifying beet coracoid and application of specific primer pair
A primer pair, sugar beet technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of non-specific PCR products, difficult molecular identification, unstable results, etc. Relevant knowledge requirements, easy to carry out widely, easy to operate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Example 1 sample DNA extraction
[0020] Genomic DNA extraction of sugar beet beetle weevil and other field pests adopts the kit method, and the kit used is Ezup Column Animal Genomic DNA Extraction Kit; purchased from Sangon Bioengineering Co., Ltd. The specific operation steps are as follows:
[0021] (1) Soak the preserved insect samples in absolute ethanol, separate the single-headed insects (eggs, pupae), wash them twice with sterile water, and place them in sterile water for 24 hours to completely remove the ethanol residue. Afterwards, the worms were placed on sterilized filter paper to dry;
[0022] (2) Put the dried sample into a new 1.5mL sterilized centrifuge tube, add digestion solution and grind the sample fully with an electric grinder, then perform DNA extraction according to the DNAEzup column animal genome extraction kit operation manual;
[0023] (3) When the sample DNA is on the adsorption column in the last step, add 50 μL of sterilized deionized w...
Embodiment 2
[0025] Embodiment 2: the RAPD amplification of sample DNA
[0026] According to the principle of RAPD, 20 primers of RAPD 10-mer Set B, a random primer set from BGI, were selected for PCR screening of DNA samples of beet beetle weevil and other field pests.
[0027] Use Tiangen 2×Taq PCR MasterMix II reagent for PCR amplification reaction. The total reaction system is 25 μL, including: 1 μL of sample DNA to be tested; 1 μL of random primers; 12.5 μL of 2×Taq PCR MasterMix II; 10.5 μL of sterile water. The DNA concentration of the sample to be tested is not less than 250pg / μL; the molar concentration of the random primer is 10μM. The PCR program was: 95°C for 3min; 95°C for 30s, 40°C for 45s; 72°C for 1min, a total of 35 cycles; 72°C for 2min, and 4°C to store the PCR product. 3 μL of the PCR product was used for agarose gel electrophoresis to detect the amplification result. The agarose gel was 1.0%, the voltage used was 180V, and a gel imaging system was used to take pictur...
Embodiment 3
[0028] Example 3: Design and identification of specific primers for the sugar beet beetle beak
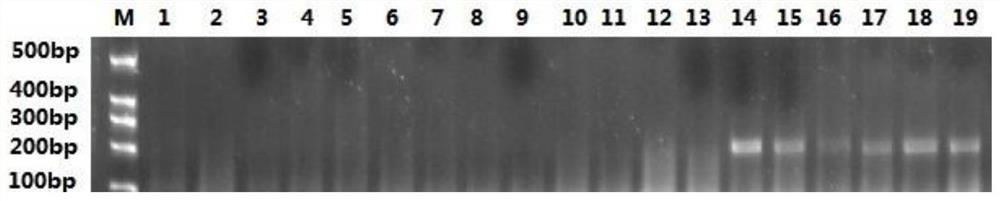
[0029] The PCR product was sequenced to obtain a definite base sequence of 428bp. After analyzing the sequence, use Primer Premier5 software to design specific primers. Upstream primer sequence: SEQ ID NO: 1: TGTCGTGAAGTCAACTACTGATG, downstream primer sequence: SEQ ID NO. 2: CGCTACTCACGATGATCTCTGATC. The primers were sent to Sangon Bioengineering Co., Ltd. for synthesis. The predicted fragment size of the PCR product of the pair of primers is 223bp.
[0030] Amplification and identification of DNA samples from different developmental stages of beet beetle weevil (eggs, early larvae, mature larvae, pupae, female adults and male adults) and DNA from other field pests were carried out. Use Tiangen 2×Taq PCR MasterMix II reagent for PCR amplification reaction, the total reaction system is 25 μL, including: 1 μL of sample DNA to be tested; 0.5 μL of upstream primers; 0.5 μL of downst...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com