Combined marker for predicting prognosis of liver cancer and application of combined marker
A liver cancer prognosis and combined biological technology, applied in the field of biomedicine, can solve problems affecting tumor cell growth and invasion, achieve high sensitivity and accuracy, shorten survival time, and improve accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1TCGA database RNA-Seq sequencing data and download of patient clinicopathological data
[0030]Download the RNA-Seq sequencing data of liver cancer tissue and normal tissue next to cancer from the TCGGA database, and download the clinicopathological data of liver cancer patients at the download website: https: / / portal.gdc.cancer.gov / . In the TCGA database, 422 patients with liver cancer had clinical pathological data; 373 patients with liver cancer had RNA-Seq sequencing data of liver cancer tissue. A total of 369 patients with liver cancer had both clinical pathological data and hepatocellular carcinoma tissue sequencing data, of which 50 patients with liver cancer had RNA-Seq sequencing data of liver cancer tissue and normal liver tissue next to cancer. Since the expression profile data of mRNA has been standardized by TCGAs, no further standardization of these data will be carried out, and the pathological parameters of patients with liver cancer are shown in Ta...
Embodiment 2
[0033] Example 2 Screening of differentially expressed gene sets in patients with liver cancer
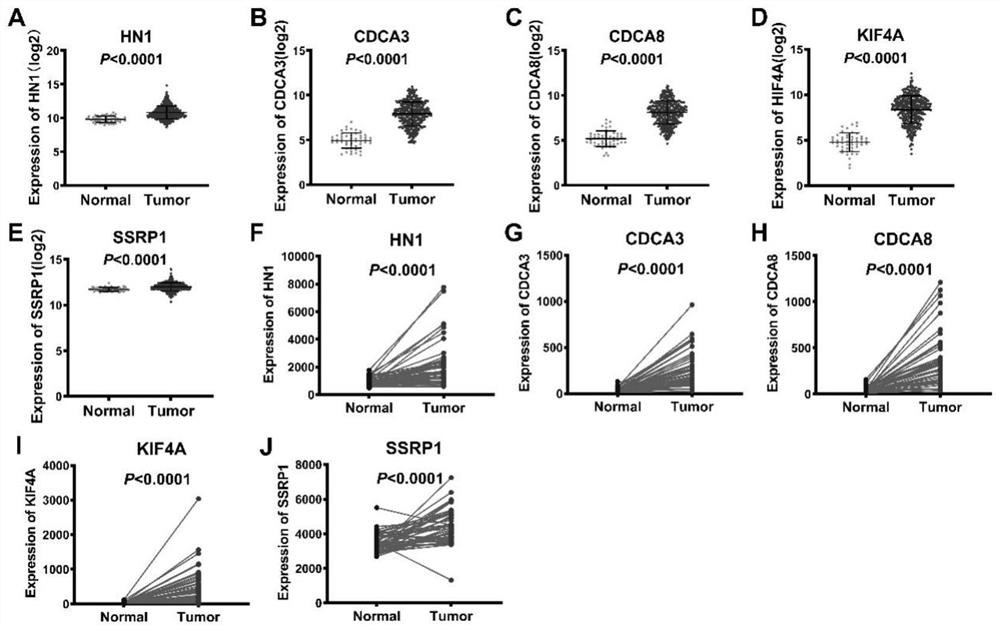
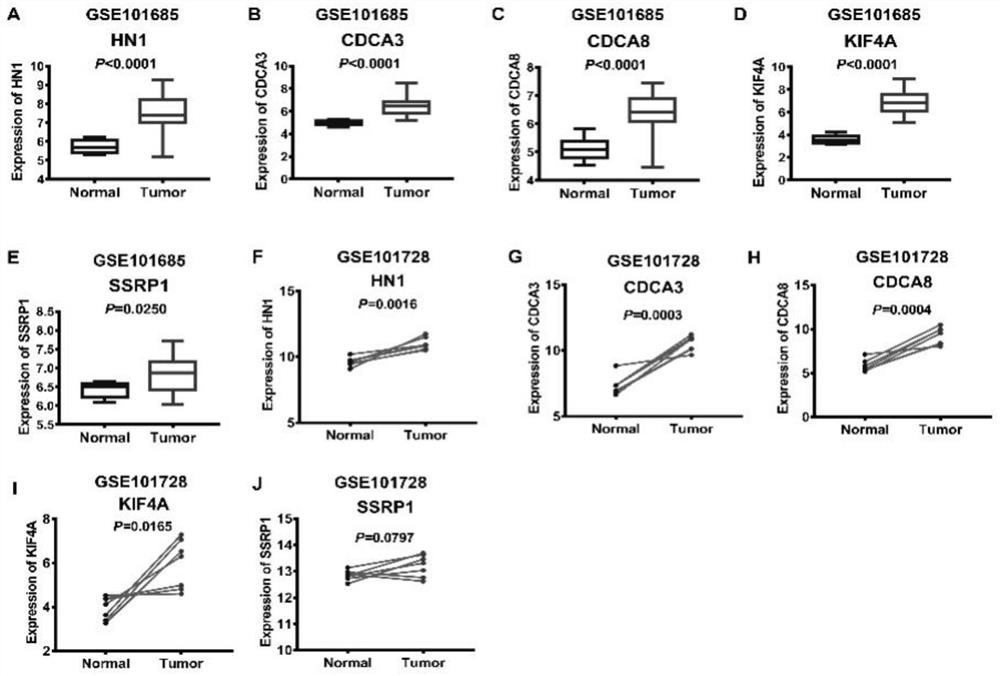
[0034] Using GSEA version 4.1.0, RNA-Seq sequencing data of liver cancer tissue and paracancerous normal liver tissue were used to analyze the differentially expressed gene sets in liver cancer tissue and paracancerous normal liver tissue. | NES|>1.5, NOM P-val<0.05 are the criteria for screening abnormally expressed gene sets in liver cancer tissues to determine gene sets that have predictive prognosis value in liver cancer treatment; | NES | represents the normalized enrichment analysis score, and NOM p-val represents the corrected p value, characterizing the credibility of the enrichment results; Among them, the transcription factor E2F gene contains 197 genes in the concentration, and its | NES | =2.071552 and NOM p-val=0.001961 are the gene sets with the largest differences among liver cancer tissues and paracancerous normal tissues (Table 2).
[0035] Table 2. Abnormally expresse...
Embodiment 3
[0037] Example 3 Construction of a prognosis model for liver cancer
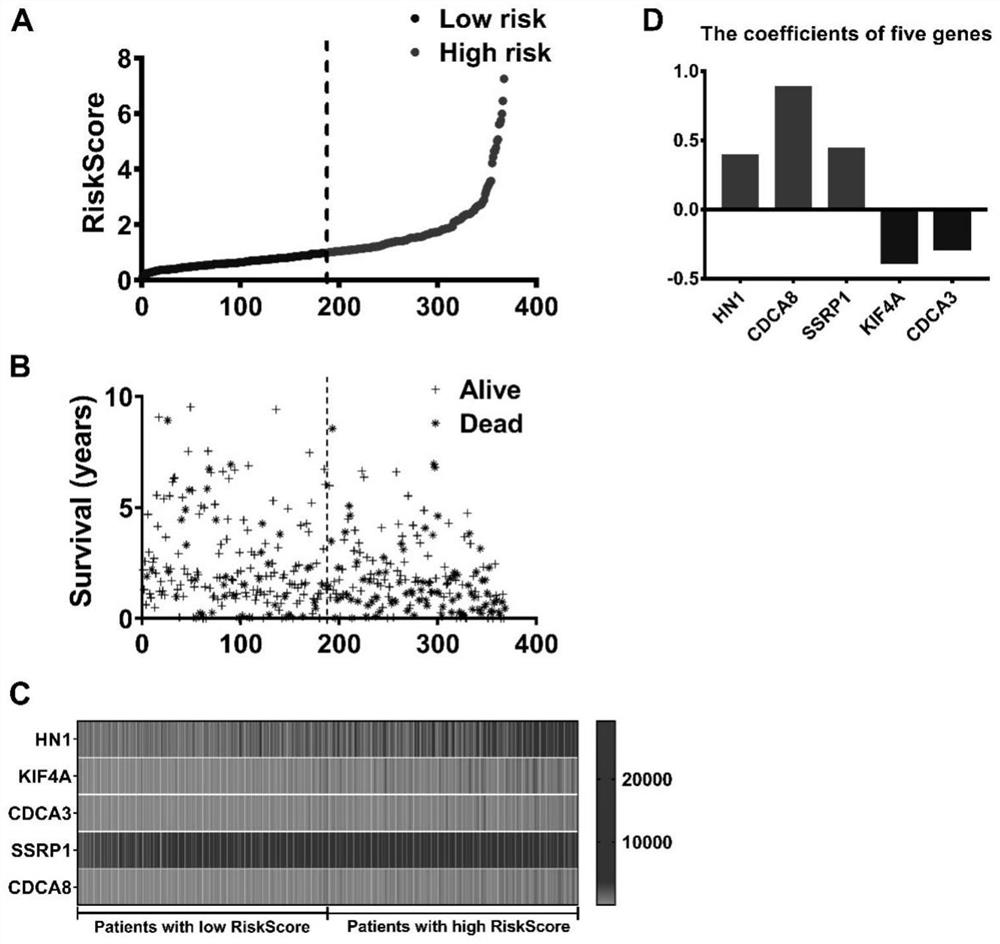
[0038]Using univariate Cox regression analysis, the differential genes screened from GSEA affected the prognosis of liver cancer patients, and the PFigure 1 A), the present invention found that the high-risk group has a low overall survival time of the low-risk group, the high number of deaths ( Figure 1 B)。
[0039] Table 3.Multivariate COX analysis results of genes in liver cancer prognosis model
[0040]
[0041]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com