Method for determining minimum entrapment binding ratio in polypeptide and siRNA co-assembly
A technology of co-assembly and binding ratio, applied in the field of bioengineering, can solve problems such as the minimum encapsulation and binding ratio of peptides and siRNA that are not recorded in experiments.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
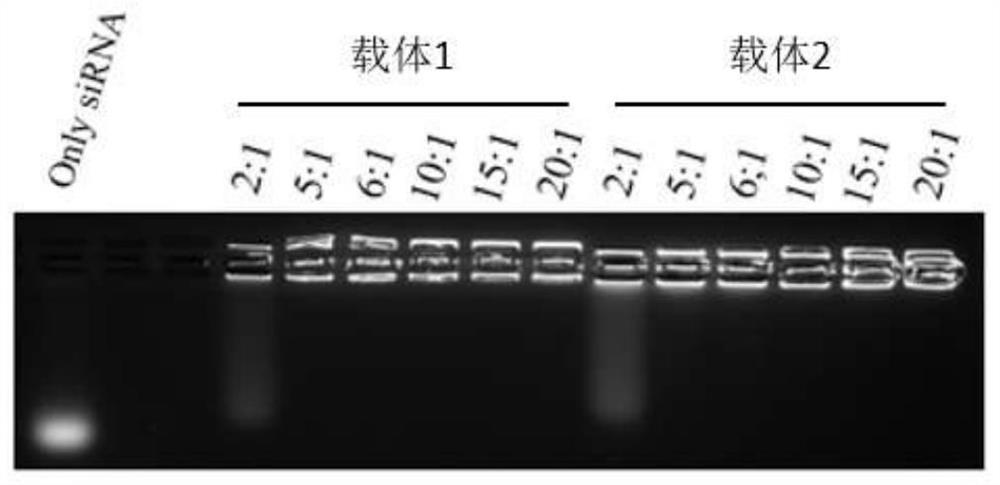
[0059] This embodiment discloses an experimental method for detecting the binding ratio of carrier polypeptide to siRNA, which specifically includes the following steps:
[0060] (1) Prepare 2% agarose gel in advance. The preparation method is as follows: a. Weigh 3.2g of agarose powder into a beaker, add 1×TAE to make the volume to 160mL, shake and mix slightly, and place it in a microwave oven to heat for 4- 7min, until completely dissolved, after cooling for a while, add GelRed Nucleic Acid Gel Stain (BIOTIUM; Cat: 41003) dye to make the working concentration 1×; b. Pour the dissolved agarose into the mold inserted into the comb, and leave it at room temperature for 30min, Pull out the comb; c. Put the prepared gel into the electrophoresis tank, and perform electrophoresis experiment after adding the sample.
[0061] (2) Sample preparation method:
[0062] The CPP and siRNA were configured in a series of ratios as shown in Table 1 below. Polypeptide stock solution concent...
Embodiment 2
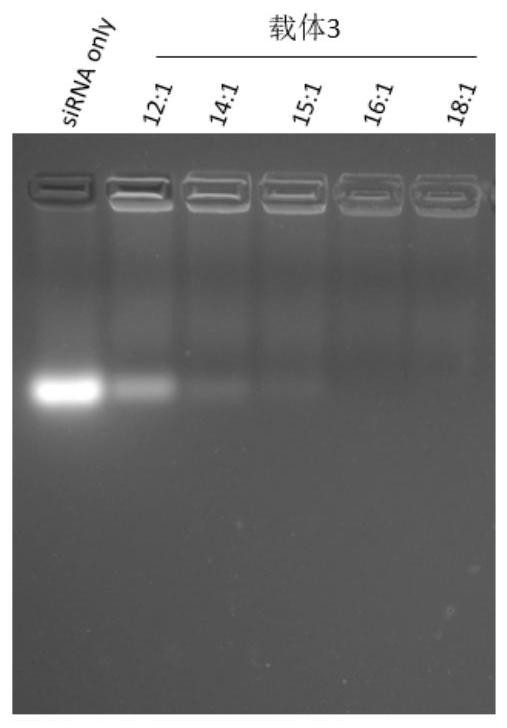
[0075] This embodiment discloses an experimental method for detecting the binding ratio of carrier polypeptide to siRNA, which specifically includes the following steps:
[0076] (1) Prepare 2% agarose gel in advance. The preparation method is as follows: a. Weigh 3.2g of agarose powder into a beaker, add 1×TAE to make up to 160ml, shake and mix evenly, and heat in a microwave oven for 4- 7min, until completely dissolved; b. Pour the dissolved agarose into the mold inserted into the comb, leave it at room temperature for 30min, and pull out the comb; c. Put the gel into the electrophoresis tank, and conduct electrophoresis experiment after adding the sample .
[0077] (2) Sample preparation method:
[0078] The CPP and siRNA were configured in a series of ratios as shown in Table 3 below. Polypeptide stock solution concentration: 500 μM; siRNA stock solution concentration: 50 μM solution.
[0079] table 3
[0080]
[0081] The CPP and siRNA were configured in a series o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com