Homologous recombination defect detection method
A defect detection and homologous recombination technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as insufficiency, inability, and large amount of data
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1: Total DNA extraction
[0041] For a puncture sample, use the QIAamp DNA FFPE Tissue Kit (QIAGEN, Cat. No. 56404) to extract the FFPE paraffin sample. After extraction, 25ng-200ng will be obtained, of which 200ng will be taken out for DNA fragmentation.
Embodiment 2
[0042] Example 2: DNA Fragmentation
[0043] 1. Set up Covaris S220
[0044] Pour fresh deionized water into the sink until level = 12 (Covaris S220). Turn on the power switches of the Covaris, the cooling unit and the circulation system in sequence. If the loop system front panel shows OFF, press and hold the Enter key to turn the switch on. Set the cooling device to 4°C. Deionized water needs to be vented for at least 30 minutes. Keep the pump on at all times during sample processing.
[0045] 2. Prepare the samples that need to be interrupted
[0046] In a 1.5mL low adsorption tube, dilute different concentrations of sample gDNA or FFPE DNA to a volume of 52.5μL with 1x Low TE, taking the maximum value in the table below, vortex to mix, and centrifuge briefly.
[0047] DNA concentration DNA Quality Rating Total Input (ng) DNA concentration ≥ 1.2 A 65-220 DNA concentration ≥ 1.2 B 65-330 DNA concentration ≥ 2.5 C 130-440
[0048] ...
Embodiment 3
[0065] Example 3: Joint Connection
[0066] 1. Connector connection
[0067] Take out the reagents Ligation Buffer, Ligation Enzymes and UDIadapters from the kit in the -20°C refrigerator. The Buffer and adapter are thawed at room temperature, vortexed and mixed, then centrifuged briefly and placed on ice. After the enzyme is flicked, centrifuged briefly and placed on ice. Note that different indexes are selected for libraries built in the same batch.
[0068] Prepare Ligation's Master Mix in a 1.5 mL sterile tube and operate on ice.
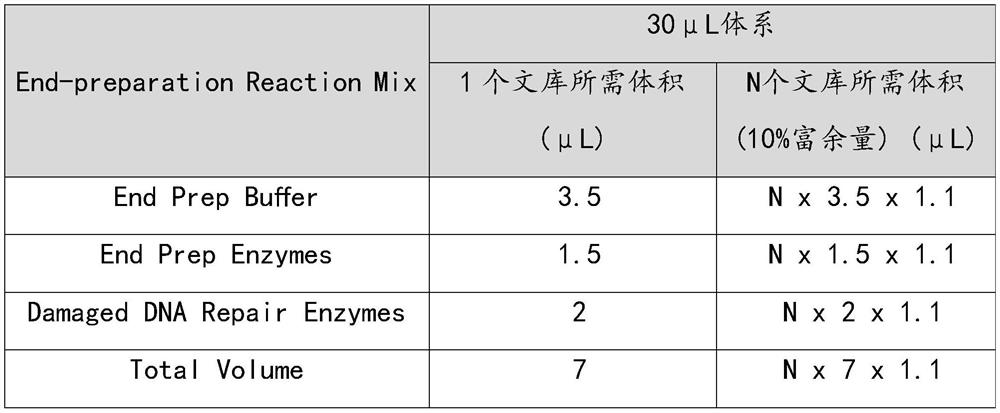
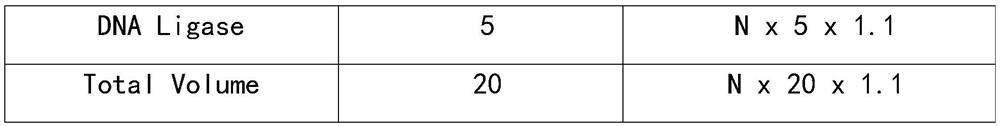
[0069]
[0070]
[0071] Set the program on the PCR machine in advance, 60min at 20℃, hold at 4℃, named Ligation.
[0072] NOTE: The heated lid temperature is set to 103°C.
[0073] 20℃ 60min 4℃ Hold
[0074] To each end-repaired 0.2 mL PCR tube, add 20 μL of Ligation Master mix. Add 5 μL of the corresponding Adapter to each tube, mix by slowly pipetting 10 times with a pipette, and centrifuge briefly until the fi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com