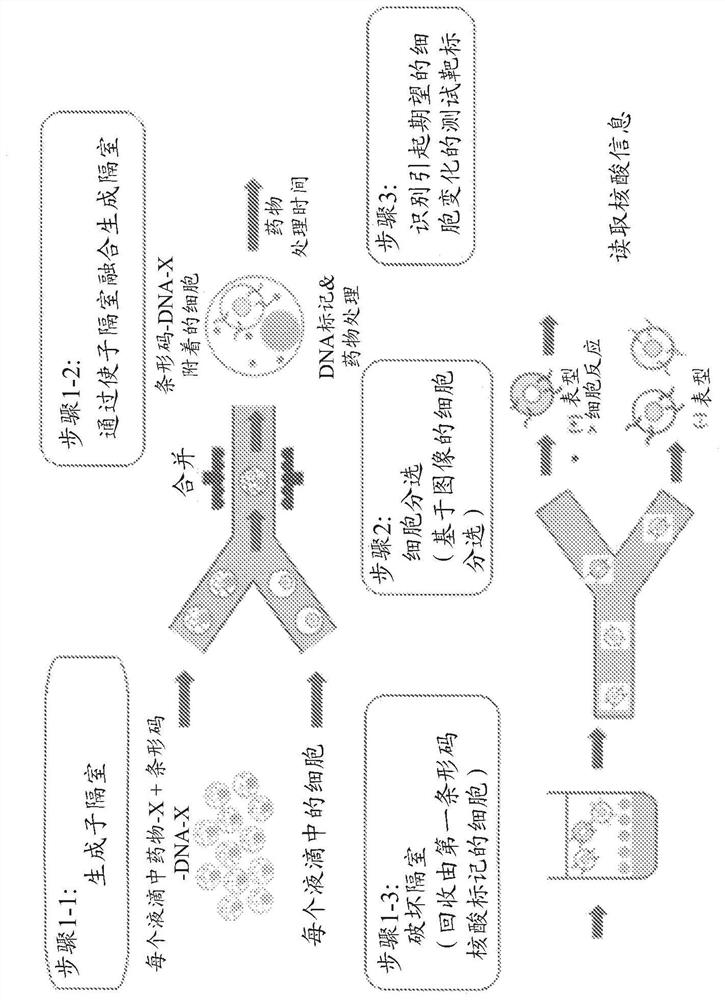
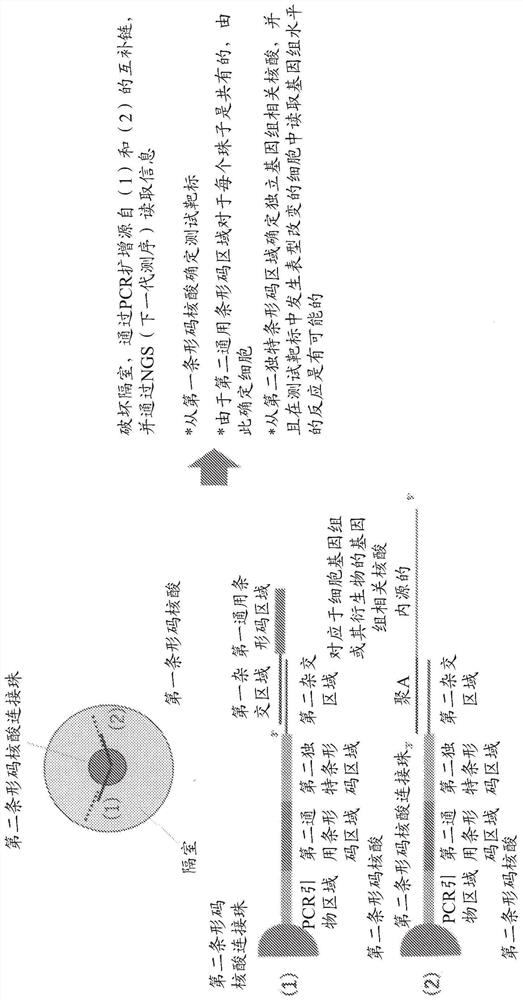
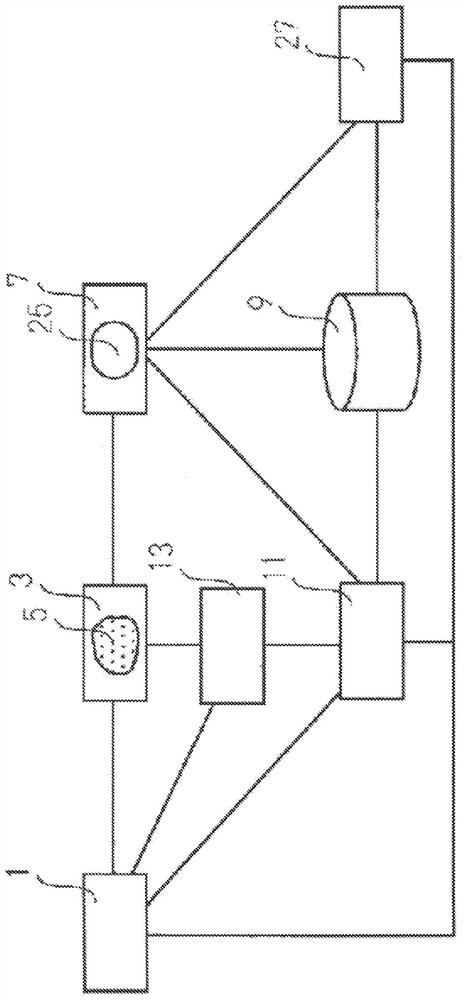
Novel cell phenotype screening method
A cell and phenotype technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of difficult gene expression reaction, mechanism analysis, slow speed, high cost, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0219] Hereinafter, the present application will be specifically described based on examples, but the present application is not limited to these examples. In addition, unless otherwise specified, the measurement methods and units of this application conform to the Japanese Industrial Standards (JIS).
reference example 1
[0220] Reference Example 1: Preliminary test for the connection of the first barcoded nucleic acid to cells
[0221] According to the Multi-seq method (described in Nature Methods, Vol. 16, pp. 619-626, (2019)), the same cells, anchor CMOs, co-anchor CMOs and oligonucleotides were used as in Example 1 described below The acid was subjected to the following preliminary tests. That is, cells and anchor CMO were incubated in phosphate-buffered saline (PBS) solution at 4°C for 5 minutes, then co-anchor CMO was added thereto and further incubated at 4°C for 5 minutes, and finally, the red fluorescent A dye (Cy5) conjugated oligonucleotide (having a sequence corresponding to a partial sequence of the first barcode nucleic acid) was mixed with and incubated at 4°C for 5 minutes.
[0222] result as Figure 4 As shown in A and B in PBS solution, red fluorescent dye (Cy5)-conjugated oligonucleotides (with a sequence corresponding to the partial sequence of the first barcode nucleic ...
reference example 2
[0223] Reference Example 2: Preliminary test for the connection of the first barcoded nucleic acid to cells
[0224] Additionally, in addition to incubation with cell culture medium containing serum or bovine serum albumin (BSA) as solvent, use Test This preliminary experiment was carried out in the same manner as in Example 1. As a result, it was confirmed that the red fluorescent dye (Cy5)-conjugated oligonucleotide (having a sequence corresponding to the partial sequence of the first barcode nucleic acid) had a reduced rate of attachment to cells, such as Figure 4 Photos in C and D are shown.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com