Composition, kit and method for detecting genome DNA of Vero cell
A technology of composition and kit, which is applied in the detection of Vero cell genomic DNA, the composition of detection of Vero cell genomic DNA, and the field of composition, which can solve the problem that the detection sensitivity is not high enough, the interference cannot be well eliminated, and the detection sensitivity is getting higher and higher. standards and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
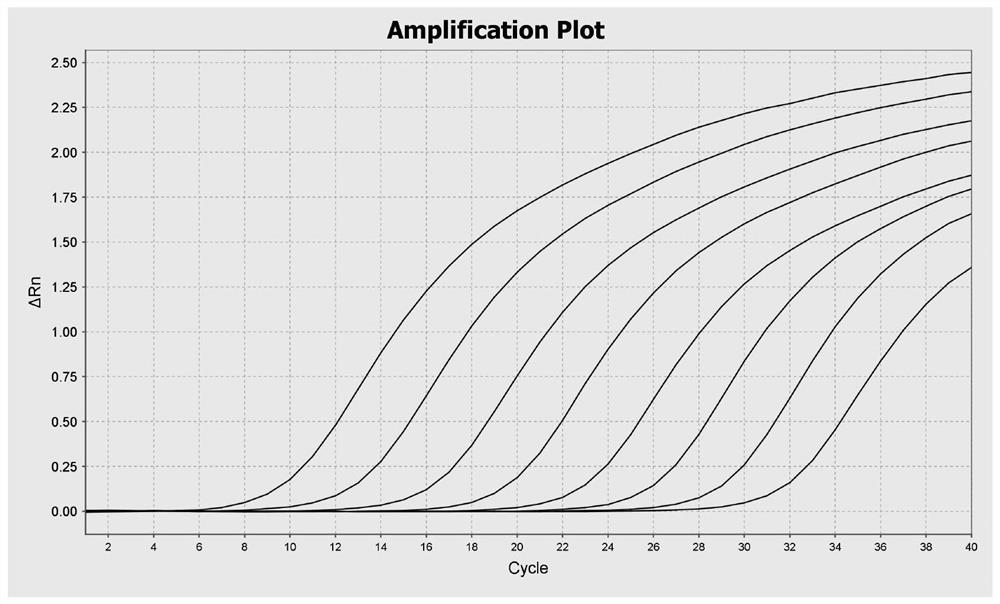
[0095] (2) Preparation of standard DNA
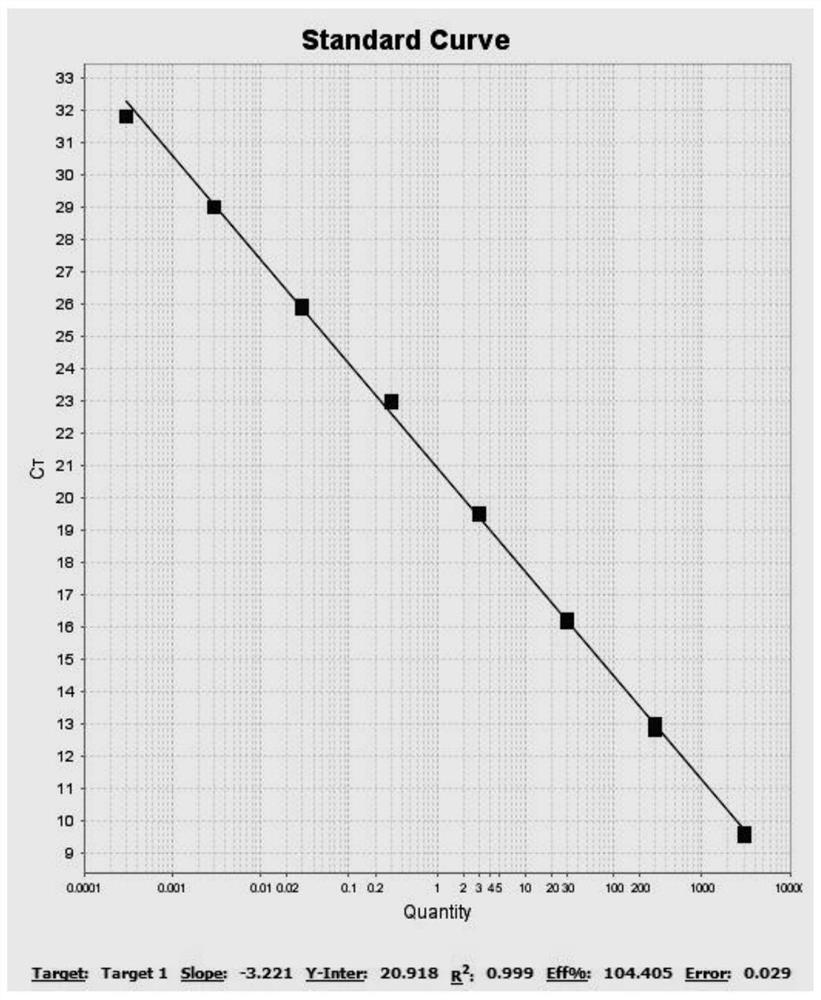
[0096] The Vero cell genomic DNA purchased from the China Institute of Testing and Inspection was diluted with nucleic acid standard dilution (Tiangen Biochemical Products, catalog number RT504), and the diluted concentration was shown in Table 2, wherein the non-template control (NTC) for ultrapure water.
[0097] Table 2:
[0098] Gradient number 1 2 3 4 5 6 7 8 concentration 300pg / μL 30pg / μL 3pg / μL 300fg / μL 30fg / μL 3fg / μL 0.3fg / μL 0.03fg / μL
[0099] (3) Probe method qPCR reaction system
[0100] The reaction system is shown in Table 3:
[0101] table 3:
[0102] component Volume required for a single reaction (μL) Template DNA (standard DNA or ultrapure water) 10 2×qPCR reaction reagents 15 Upstream primer (10μM) 0.75 Downstream primer (10μM) 0.75 Detection Probe (10μM) 0.6 Ultra-pure water 2.9 total capacity 30
[0103] (4) Prob...
Embodiment 1
[0113] Example 1 Primer, probe design and sensitivity test
[0114] 1.1 Obtaining the polynucleotide sequences of primers and probes for designing and detecting Vero cell genomic DNA:
[0115] (1) The sequence file GCF_015252025.1_Vero_WHO_p1.0_genomic.fna.gz and the repeat region annotation file GCF_015252025.1_Vero_WHO_p1.0_rm.out.gz were downloaded from the NCBI database to obtain the genome of the African green monkey (Chlorocebus sabaeus). The annotation files in the repeat area were obtained by using the RepeatMasker program (version: open-4.0.8), the RepBse database (20181026) and the Dfam_Consensus (20181026) database.
[0116] (2) According to the repeat annotation file information and the characteristics of different repeat sequences and their distribution characteristics in the genome, on the corresponding genome, extract the AluSz and AluY repeat sequences of the SINE / Alu family, and extract the satellite sequence ALR / Alpha type repeat sequences, and filtered to r...
Embodiment 2
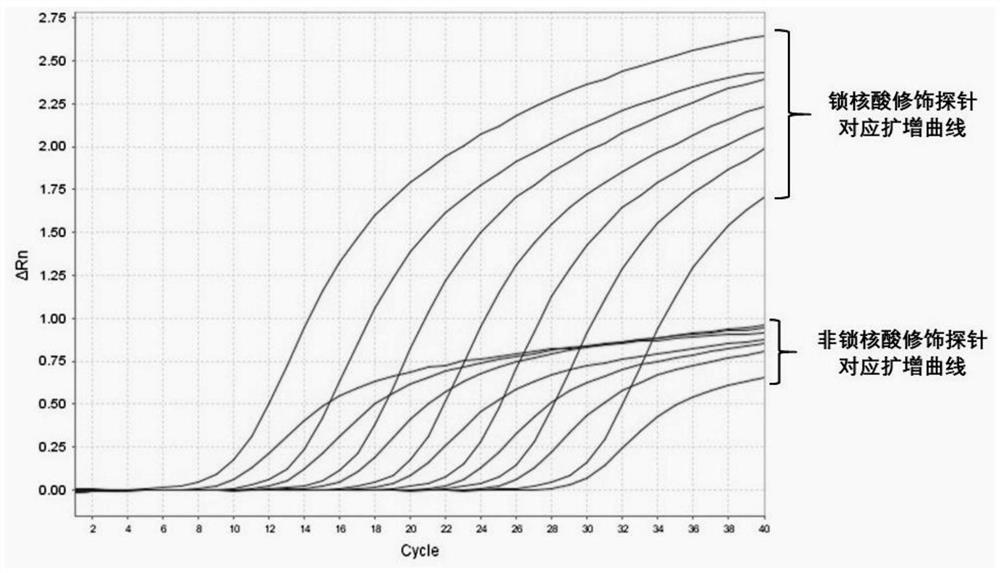
[0139] Example 2 Influence of probe-locked nucleic acid modification on detection results
[0140] The primer probes used in this example are as follows:
[0141] Upstream primer: CATCTCACAGAGTTACATCTTTCC (SEQ ID NO: 2);
[0142] Downstream primer: CTTTTTCACCATAGCCCTCTA (SEQ ID NO: 3);
[0143] Detection probe: CC T TTCGCTAAGGCTGTTCTTGT (SEQ ID NO: 4), the underlined base is modified by locked nucleic acid;
[0144] Detection probe (unlocked nucleic acid modification): CCTTTCGCTAAGGCTGTTCTTGT;
[0145] Amplification product: CATCTCACAGAGTTACATCTTTCCCTTCAAGAAGCCTTTCGCTAAGGCTGTTCTTGTGGAATTGGCAAAGTGATATTTGGAAGCCCATAGAGGGCTATGGTGAAAAAG (SEQ ID NO: 5).
[0146] The qPCR system is the same as in Example Table 2:
[0147] The DNA concentration settings of the standards used in the qPCR standard curve are shown in Table 8:
[0148] Table 8:
[0149] gradient 1 2 3 4 5 6 7 concentration 300pg / μL 30pg / μL 3pg / μL 300fg / μL 30fg / μL 3fg / μL 0.3fg / μL ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com