Primer and method for molecular identification of channel catfish and ietalurus punetaus
A technology for channel catfish and cloud catfish, applied in the field of molecular biology, can solve problems such as escape, ecological damage in water areas, difficult identification, etc., and achieve the effects of high repetition rate, good stability, and wide application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
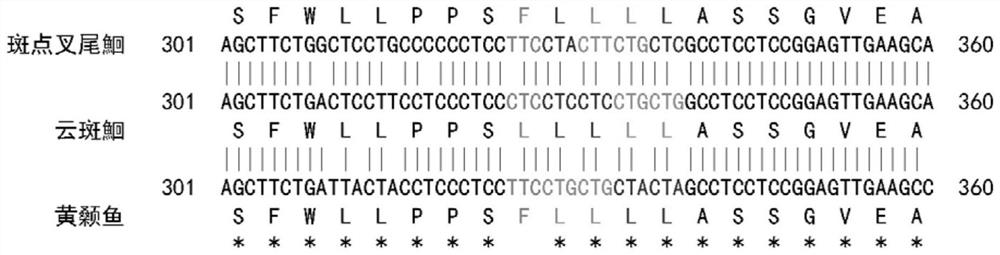
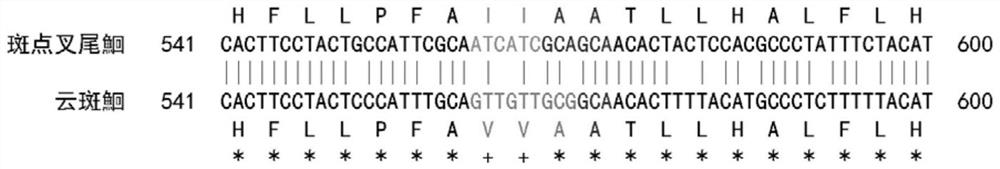
[0022] Example 1: Molecular identification of channel catfish, channel catfish and yellow catfish
[0023] 1. Primer design:
[0024] Design specific PCR amplification primer pairs, primer pair 1 sequence is: upstream primer SEQ ID NO.1: 5'-AAGACATTGGYACCCTYTAC-3', downstream primer SEQ ID NO.2: 5'-ATAGGAGGACRGCYGTRATA-3'.
[0025] 2. Sample collection
[0026] Channel catfish, channel catfish and yellow catfish to be identified, each with 30 individuals, were purchased from the market.
[0027] 3. Genomic DNA extraction
[0028] About 20 mg of caudal fin tissue was taken from each fish, and the genomic DNA of all individuals was extracted using a genome extraction kit. DNA integrity was identified by 1.5% agarose gel electrophoresis, and its OD value was measured by UV spectrophotometer, diluted to 50ng / μL, and stored at -20°C for later use.
[0029] 4. PCR amplification and detection
[0030] Using the extracted DNA as a template, PCR amplification was performed with th...
Embodiment 2
[0035] 1. Sample collection
[0036] According to the method of Example 1, the samples were expanded to 60 individuals of channel catfish, channel catfish and yellow catfish, all of which were purchased from the market.
[0037] 2. Total DNA extraction of samples
[0038] The total DNA of the fish samples to be tested was extracted using a genome extraction kit. The total DNA extraction method was the same as that in Example 1.
[0039] 3. PCR amplification and detection
[0040] The primers SEQ ID NO.1 and SEQ ID NO.2 were used for PCR amplification of the DNA to be detected and the total DNA, and the method was the same as that in Example 1. PCR products were detected and separated in 1% agarose gel electrophoresis. After electrophoresis at 120 V for 30 min, the size and purity of PCR amplification products were determined by gel imaging analysis system.
[0041] 4. Sequencing comparison
[0042] The detected PCR amplification product was directly sent to the biological...
Embodiment 3
[0044] Example 3 Molecular identification of channel catfish and channel catfish
[0045] 1. Primer design
[0046] A pair of specific PCR amplification primers were designed, the primer sequences are: upstream primer SEQ ID NO.3: 5'-AACCCGATTCTTCGCATTYC-3', downstream primer SEQ ID NO.4: 5'-CCGATGATGATRAATGGGTG-3'.
[0047] 2. Sample collection
[0048] Channel catfish, channel catfish and yellow catfish to be identified, 30 individuals each, were purchased from the market.
[0049] 3. Genomic DNA extraction
[0050] About 20 mg of caudal fin tissue was taken from each fish, and the genomic DNA of all individuals was extracted using a genome extraction kit. DNA integrity was identified by 1.5% agarose gel electrophoresis, and its OD value was measured by UV spectrophotometer, diluted to 50ng / μL, and stored at -20°C for later use.
[0051] 4. PCR amplification and detection
[0052] Using the extracted DNA as a template, PCR amplification was performed with the above prim...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com