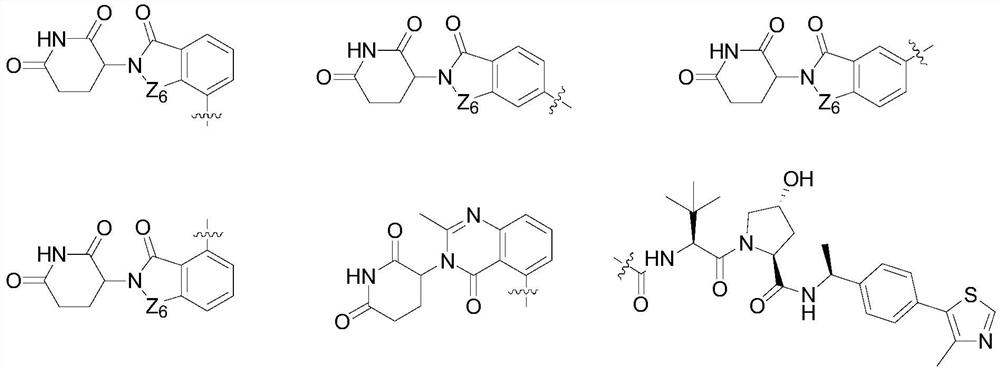
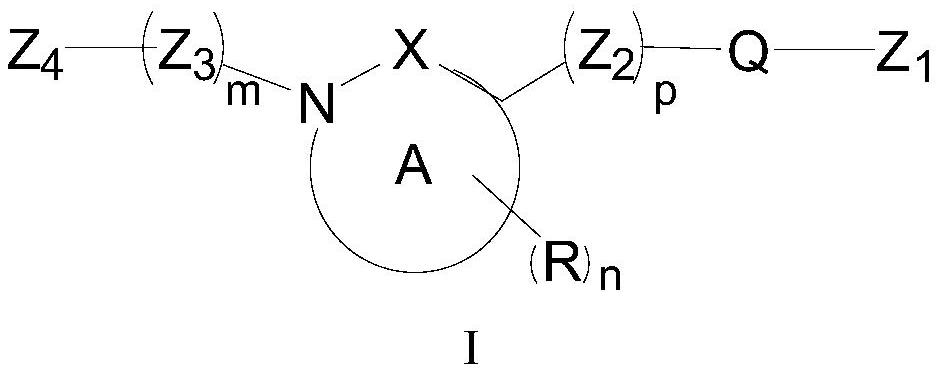
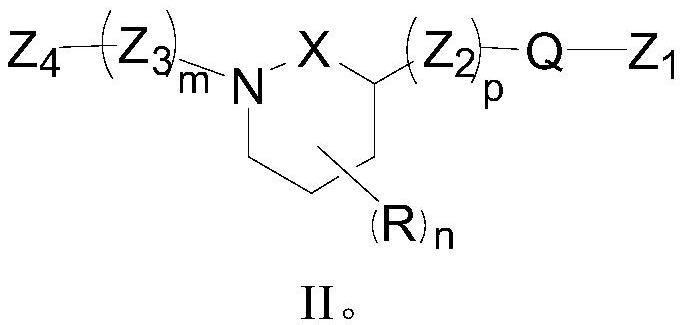
Difunctional MDM2 protein degradation agent as well as preparation method, pharmaceutical composition and application thereof
A technology of protein degradation and MDM2, which is applied in the direction of drug combination, antidote, antipyretic, etc., can solve the problems of decreased apoptosis function and cell cycle arrest, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0135]2-((3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-3-methyl-1-((2S)-3-methyl-1- Synthesis of (propyl-2-sulfonamidino)butyl-2-)-2-oxopiperidin-3-yl)acetic acid
[0136]
[0137] Step 1: Methyl 4-(3-chlorophenyl)-5-(4-chlorophenyl)-5-hydroxy-2-methylpentanoate
[0138]
[0139] 4-(3-Chlorophenyl)-5-(4-chlorophenyl)-2-methyl-5-oxopentanoic acid methyl ester (36.5g) was dissolved in ethanol (300ml), cooled to 0~ 5°C, add NaBH in batches 4 (2.85g), react at 0~10 ℃ for 2 hours, TLC tracking reaction is basically complete, add acetic acid (~8ml) dropwise until no hydrogen is released, concentrate the solvent, add 300ml of ethyl acetate, wash with water and saturated sodium bicarbonate successively, After drying over anhydrous magnesium sulfate, it was concentrated to obtain 37 g of methyl 4-(3-chlorophenyl)-5-(4-chlorophenyl)-5-hydroxy-2-methylpentanoate.
[0140] Step 2: 4-(3-Chlorophenyl)-5-(4-chlorophenyl)-5-hydroxy-2-methylpentanoic acid
[0141]
[0142] Diss...
Embodiment 2
[0185] Compound 4-(2-((3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-3-methyl-1-((S)-3-methyl yl-1-((S)-2-propylsulfonamidino)butan-2-yl)-2-oxopiperidin-3-yl)acetamido)benzoic acid
[0186]
[0187] Step 1: 4-(2-((3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-3-methyl-1-((2S)-3 -Methyl-1-(N-(2,2,2-trifluoroacetyl)-2-propylsulfonamidino)butyl-2-)-2-oxopiperidin-3-yl)acetamide Synthesis of methyl) benzoate
[0188]
[0189]2-((3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-3-methyl-1-((2S)- 3-Methyl-1-(N-(2,2,2-trifluoroacetyl)-2-propylsulfonamidino)butyl-2-)-2-oxopiperidin-3-yl)acetic acid (800mg), methyl p-aminobenzoate (219mg), DMAP (324mg) and EDC.HCl (508.4mg) were successively added to the DCM (16ml) solution and stirred at room temperature overnight. The reaction was complete as detected by TLC. Water was added dropwise to the reaction solution under ice bath to quench the reaction, the cold 1N HCl solution was adjusted to pH=2, the layers were separated, ...
Embodiment 3
[0195] 4-(2-((3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-3-methyl-1-((2S)-3-methyl -1-(Propan-2-ylthiamidinyl)butan-2-yl)-2-oxopiperidin-3-yl)acetamido)-N-(5-(2-(2,6- Synthesis of Dioxopiperidin-3-yl)-1-oxoisoindol-4-yl)pent-4-yn-1-yl)-2-methoxybenzamide
[0196]
[0197] step 1:
[0198]
[0199] 3-Bromo-2-bromomethyl-benzoic acid methyl ester (5.00 g, 16.2 mmol) and 3-amino-piperidine-2,6-dione hydrochloride (2.94 g, 17.9 mmol) were added to acetonitrile (50 ml) ), then added triethylamine (8.21g, 81.1mmol), heated to reflux overnight, the reaction solution turned dark purple, cooled to room temperature and filtered, the solid was washed with water (20ml) and MTBE (20ml), and dried in vacuo to give 2.70 g solid, 52% yield. 1 H NMR (400MHz, DMSO-d 6 )δ11.03(s, 1H), 7.88(d, J=7.9Hz, 1H), 7.78(d, J=7.5Hz, 1H), 7.52(t, J=7.7Hz, 1H), 5.16(dd, J=13.3, 5.1Hz, 1H), 4.43 (d, J=17.6Hz, 1H), 4.27 (d, J=17.6Hz, 1H), 2.92 (ddd, J=18.1, 13.6, 5.4Hz, 1H), 2.65–2.51 (m, 1H)...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com