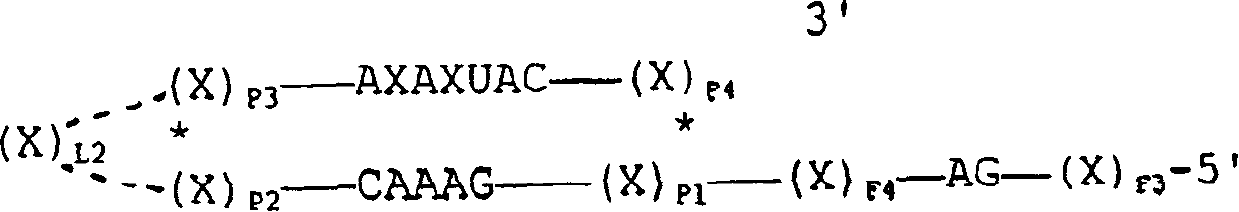Ribozymes targeting retroviral packaging sequence expression constructs and recombinant retroviruses containing such constructs
A sequence and ribozyme technology, applied in the direction of medical preparations containing active ingredients, medical raw materials derived from viruses/phages, antiviral agents, etc., can solve short serum half-life, rapid bile clearance rate, poor oral utilization, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0141] Cleavage of Mo-MLV Psi Packaging Sequence Catalyzed by Ribozyme in Vitro
[0142] To show that the target site is indeed cleavable, an in vitro cleavage reaction was performed in cell culture medium prior to detection of the ribozyme.
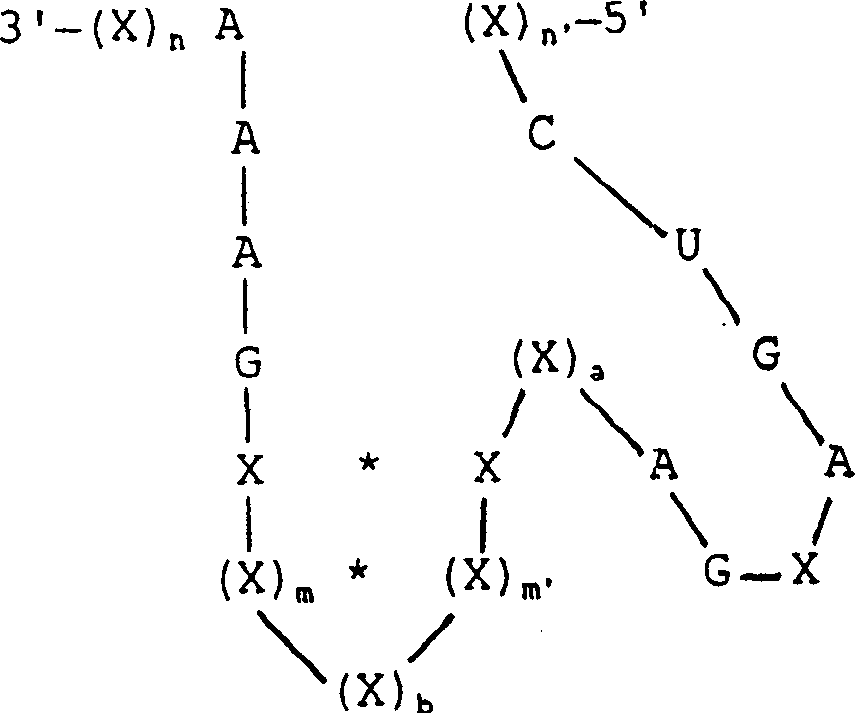
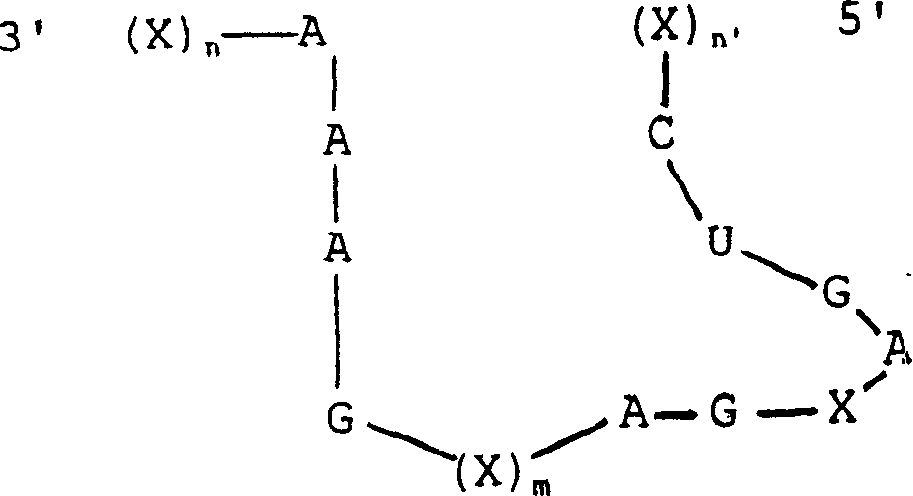
[0143]Four sites were selected in the Mo-MLV packaging region based on the presence of GUC bases and the potential accessibility of sites within the RNA secondary structure suggested from Zuker's FOLDRNA program (Zuker et al., 1981). These sites were named 243, 274, 366 and 553 based on the nucleotide distance from the 5' end of the viral transcript (see Figure 2). These nucleotide positions are described in RNA tumor viruses (Coffin, 1985). Two types of ribozymes were designed: three mononucleases targeting positions 243, 274, and 366, respectively, with an arm-length range of 12 nucleotides, and one multinuclease targeting all four sites Enzymes with indirect arms of sequence length between each target site. The sites and overall de...
Embodiment 2
[0149] Anti-Mo-MLV packaging site (Psi) construct
[0150] After exhibiting efficient in vitro cleavage, the engineered ribozyme along with a long antisense sequence complementary to the Psi packaging region was cloned into neo r 3' untranslated region of the gene (Figure 4). neo r is a prokaryotic gene encoding a phosphorylase and thereby inactivates neomycin or the neomycin analogue G418. The latter is toxic to mammalian cells, exogenous neo r The expression of the gene allows the cell to survive. with neo r The construct of the gene-associated SV40 promoter is within the mammalian expression vector pSV2neo and is shown schematically in FIG. 4 .
[0151] The ribozyme insert and antisense control were cloned into neo by blunt end ligation r Sma I site in the 3' untranslated region. The resulting vectors were designated pSV243, pSV274, pSV366, pSVM7 and pSVasPsi (antisense construct), respectively.
Embodiment 3
[0153] Transfection of the construct into the 3T3-Mo-MLV producing cell line
[0154] Various pSV2neo-based constructs were transfected into 3T3-Mo-MLV cells using the calcium phosphate transfection method (Chen et al., 1987). Positive colonies were those formed after 9-12 days of growth in the presence of 500 μg / ml G418. For each construct, 4-7 colonies were isolated using cloning cylinders. These colonies were grown, stored in liquid nitrogen, and used for further testing. After 10-14 days of selection in 500 μg / ml G418, several stable clonal cell lines were established for each construct. To determine the integration of the transfected DNA expression constructs, genomic DNA was prepared from specifically transfected cell lines and then subjected to Southern analysis. Genomic DNA was digested with restriction enzymes Hind III and Nru I to generate neo r Fragment with insert (ribozyme or antisense sequence). then use neo r Specific probes confirm the presence of the con...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com