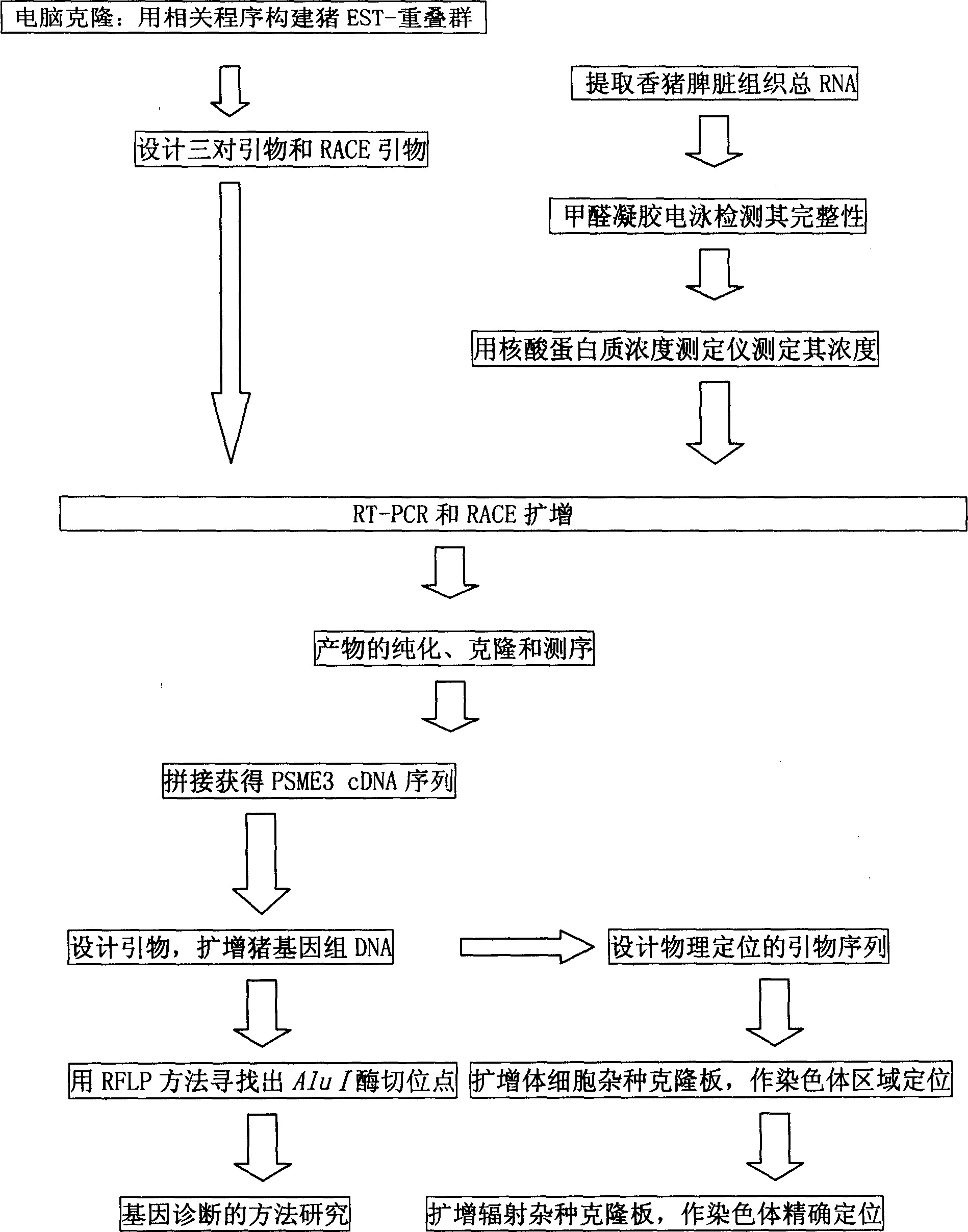
Gene PSME3 of fat thickness at back of pig as well as preparation method
A technology for pig backfat thickness and genetics, applied in the field of genetic engineering, can solve problems such as research gaps, low efficiency, and neglect of gene interactions.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0134] Example 1: Association analysis of pig marker traits
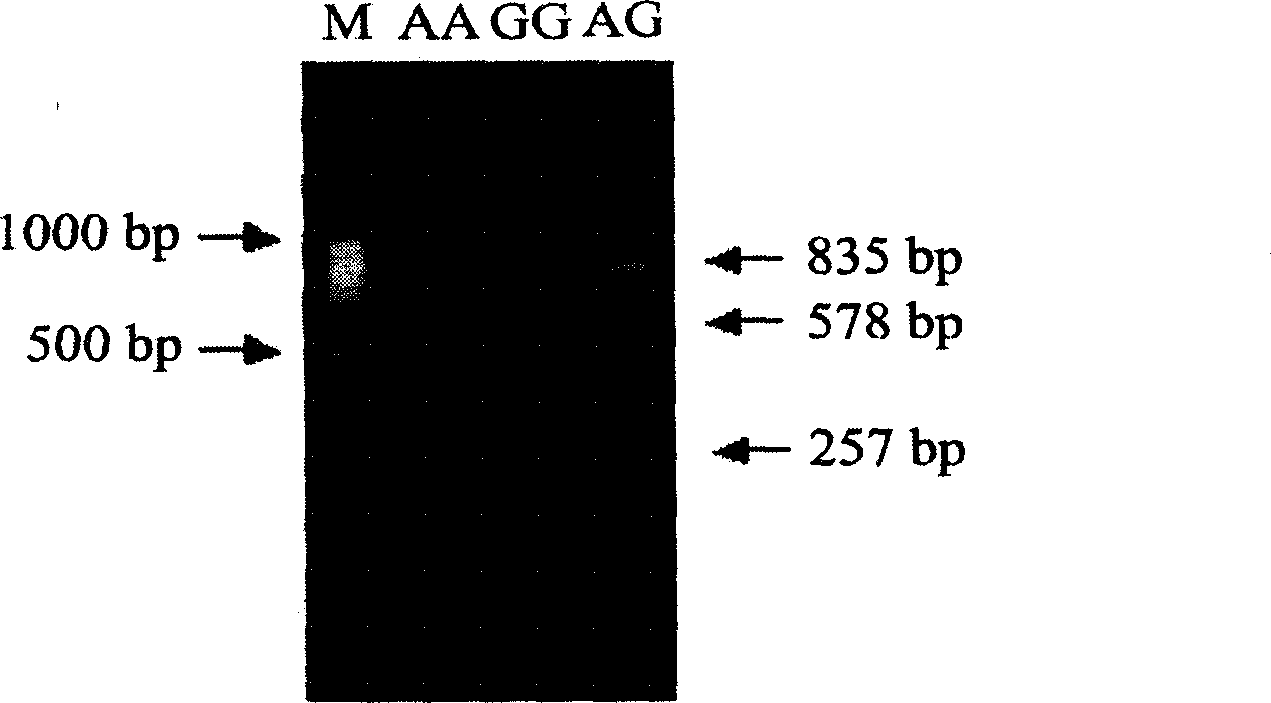
[0135] In a commercial group of large white pigs, the association analysis of the AluI-RFLP polymorphism site in exon 8 of PSME3 gene and some production traits was carried out. The traits analyzed were birth weight, weaning weight, and background fat thick.
[0136] The results of genotype detection showed that the AA genotype accounted for the vast majority of the 201 individuals, with 182 individuals, and 18 individuals with the AB genotype. The results of the simple mean and standard deviation analysis of traits between the AA and AB genotypes were summarized in Table 5, the analysis results show that there is a significant difference in the backfat thickness between the AA and AB genotypes at 170 days of slaughter (P<0.05), and there is a correlation trend between this locus and the birth weight (P=0.11).
[0137] Table 5: Association analysis of different PSME3 gene AluI-RFLP genotypes and some production ...
Embodiment 2
[0145] Example 2: Association analysis of pig marker traits
[0146] In a Tongcheng pig herd, the association analysis between the AluI-RFLP polymorphism of exon 8 of the PSME3 gene and some production traits was carried out. The traits analyzed were birth weight and backfat thickness when slaughtered up to 90 kg. The analysis results showed that the backfat thickness of BB and AB genotypes was significantly different (P<0.05) (Table 6)
[0147] Table 6: Association analysis of different PSME3 gene AluI-RFLP genotypes and some production traits
[0148] Genotype Number of individuals Birth weight Backfat thickness
[0149] Genotypes N Birth weight Backfat thickness 1
[0150] (kg) (mm)
[0151] BB90 1.1±0.27 12.18±2.11
[0152] AB 18 1.21±0.31 11.07±1.86
[0153] P value 0.16 0.04
[0154] P-value
Embodiment 3
[0155] Example 3: Distribution of PCR-RFLP-AluI polymorphism in various pig breeds
[0156] The PCR-RFLP-AluI polymorphism of porcine PSME3 gene was detected in 6 pig breeds, and the detection results are as described in Table 7. The data analysis in Table 7 shows that among the detected pig breeds, Duroc and Guizhou The frequencies of the dominant allele A in Xiang pigs were 0.8475 and 0.6406, respectively, while the frequencies of the dominant allele B in Erhualian, Tongcheng and Dazi pigs were 0.9688, 0.7884 and 0.6539, respectively. The frequencies of A and allele B are close, 0.4255 and 0.5745, respectively.
[0157] Table 7 Genotype and gene frequency of PSME3 gene AluI polymorphism in several pig breeds
[0158] Genotype Allele frequency
[0159] number of individuals
[0160] Variety
[0161] Genotype Allele frequency
[0162] No: of
[0163] Breeds
[0164] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com