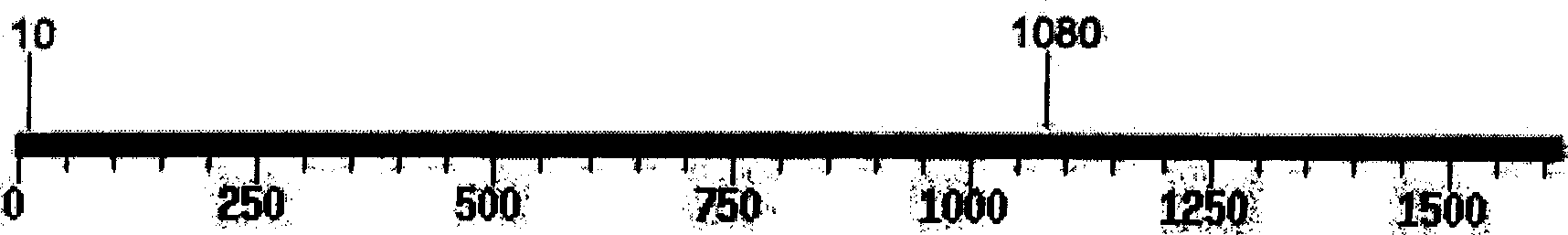Mitochondria molecular hereditary feature mark and method for identifying pure boer goat
A Boer goat, genetic marker technology, applied in sugar derivatives, organic chemistry, fermentation, etc., can solve the problems of high price, inaccuracy, and high price of purebred
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Goat Cell Mitochondrial DNA Preparation
[0063] Goat mitochondrial DNA can be prepared separately by the following three methods:
[0064] method 1.
[0065] Take a small amount of goat ear tissue or blood. Total DNA (mitochondrial DNA is included in the total DNA) was extracted from the ear tissue according to conventional methods, and dissolved with TE.
[0066] Method 2.
[0067] Add 3-10 times the volume of distilled water to the blood, treat it in boiling water for 10 minutes, centrifuge and take the supernatant as a template.
[0068] Method 3:
[0069] Animal mitochondria are first purified, and then mitochondrial DNA is extracted and purified by conventional genomic DNA extraction methods.
[0070] The DNA prepared by the above three methods can be used in the amplification reaction. Among them, method 2 is the most convenient, but the quality of template DNA is slightly worse. Method 3 is the most cumbersome, but the template DNA is the purest.
Embodiment 2
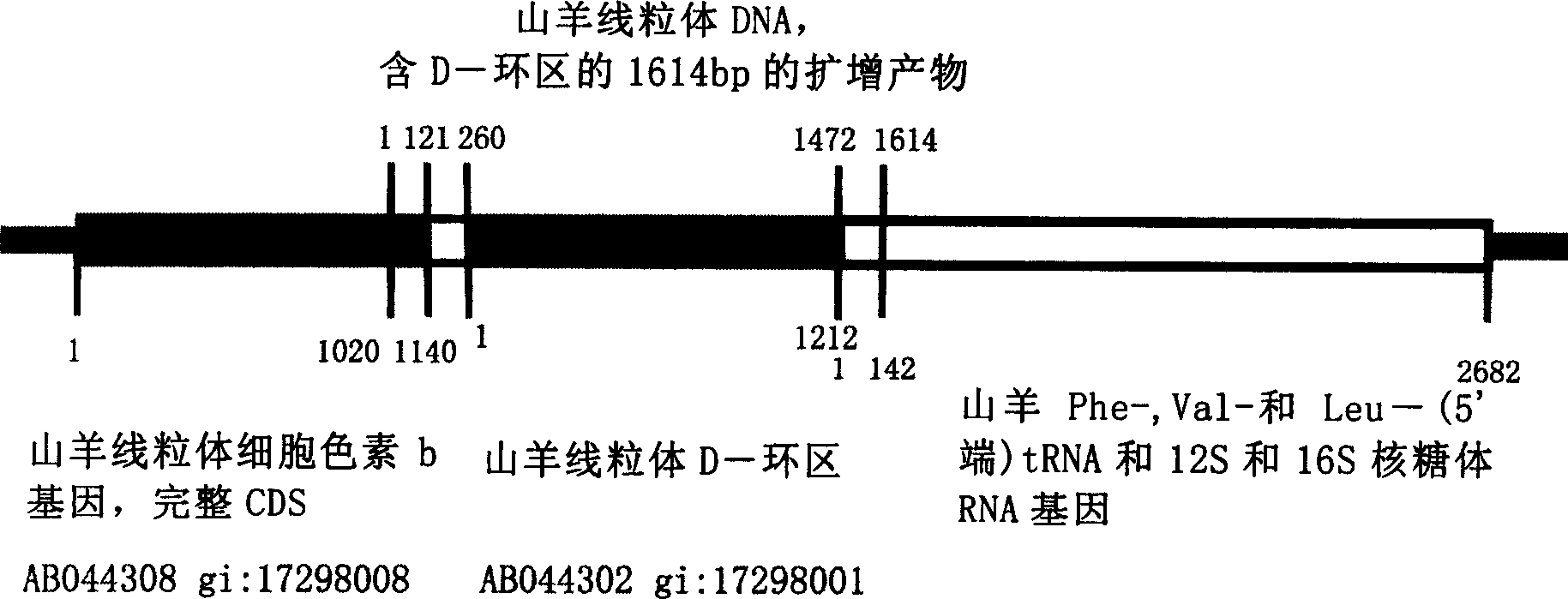
[0072] PCR Amplification of Mitochondrial D-loop Region in Goat Cells
[0073]According to Genebank goat cytochrome b gene (Capra hircus mito...[gi:17298008]) and 12srRNA gene (Capra hircus Phe-...[gi:337103]) respectively synthesize the following 3 pairs of PCR primers for three rounds of nested PCR Amplify.
[0074] Sequence (5'-3')
[0075] 1. Goatmtb528: 5' CCG ATT CTT CGC CTT CCA C 3' (SEQ ID NO: 3)
[0076] Goatmt12s783: 5'GC CCA TTT CTT CCC ATT CCA3' (SEQ ID NO: 4)
[0077] 2. Goatmtb570: 5' AGC CCT CGC CAT AGT CCA CCT 3' (SEQ ID NO: 5)
[0078] Goatmt12s699: 5'GGT TTG CTG AAG ATG GCG GTA 3' (SEQ ID NO: 6)
[0079] 3. Goatmtb1020: 5'ACA GCC AGT CGA ACA TCC CTA C 3' (SEQ ID NO: 7)
[0080] Goatmt12s142: 5'TAC TCA CCG GGG CGT GGA TG3' (SEQ ID NO: 8)
[0081] Note: b528 in the primer name indicates that it corresponds to the 528th position of the cytochrome b gene, 12s783 indicates that it corresponds to the 783rd position of the 12s rRNA gene, and so on.
[0082] T...
Embodiment 3
[0112] Restriction Enzyme Map of PCR Products in D-loop Region of Mitochondrial in Goat Cells
[0113] The Taq I restriction endonuclease from TaKaRa Company was used, and the reaction conditions recommended by the manufacturer were carried out. The total reaction volume was 20 μl. The PCR product of the third round was directly digested for 2-3 hours, and then bromophenol blue loading buffer was added to terminate the reaction. And electrophoresis in 2% agarose gel.
[0114] Results and Analysis
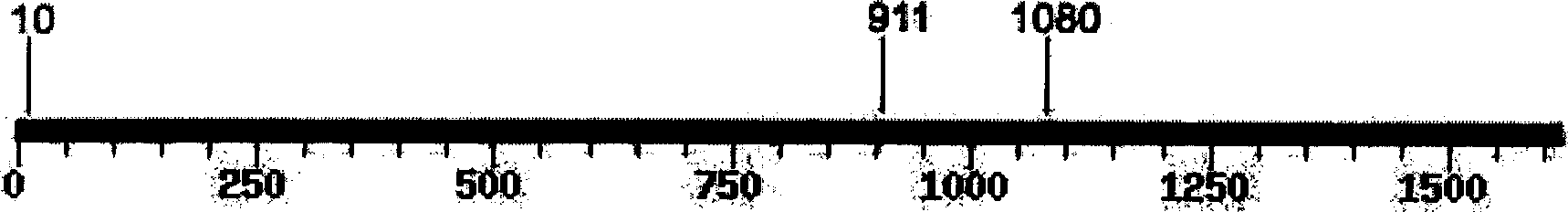
[0115] The 1614bp amplified product (SEQ ID NO: 1) of purebred Boer sheep contains an additional specific enzyme cutting site at position 911 ( Figure 2A ), so the resulting lengths were 10, 170, 533, and 901 after enzyme digestion.
[0116] The 1614bp amplified product (SEQ ID NO:2) of other goats only contains TaqI site ( Figure 2B ), so the resulting lengths were 10, 533, and 1071 after enzyme digestion. Therefore, purebred Boer sheep can be identified conveniently, quickly...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com