Use of protein fingerprint method in detecting protein fingerprint of colon cancer and rectum cancer in blood
A protein and protein technology, applied in the field of protein fingerprinting and protein fingerprinting of biological sample analysis, can solve the problems of inability to detect biomolecules, difficult to distinguish biomolecules, complex and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
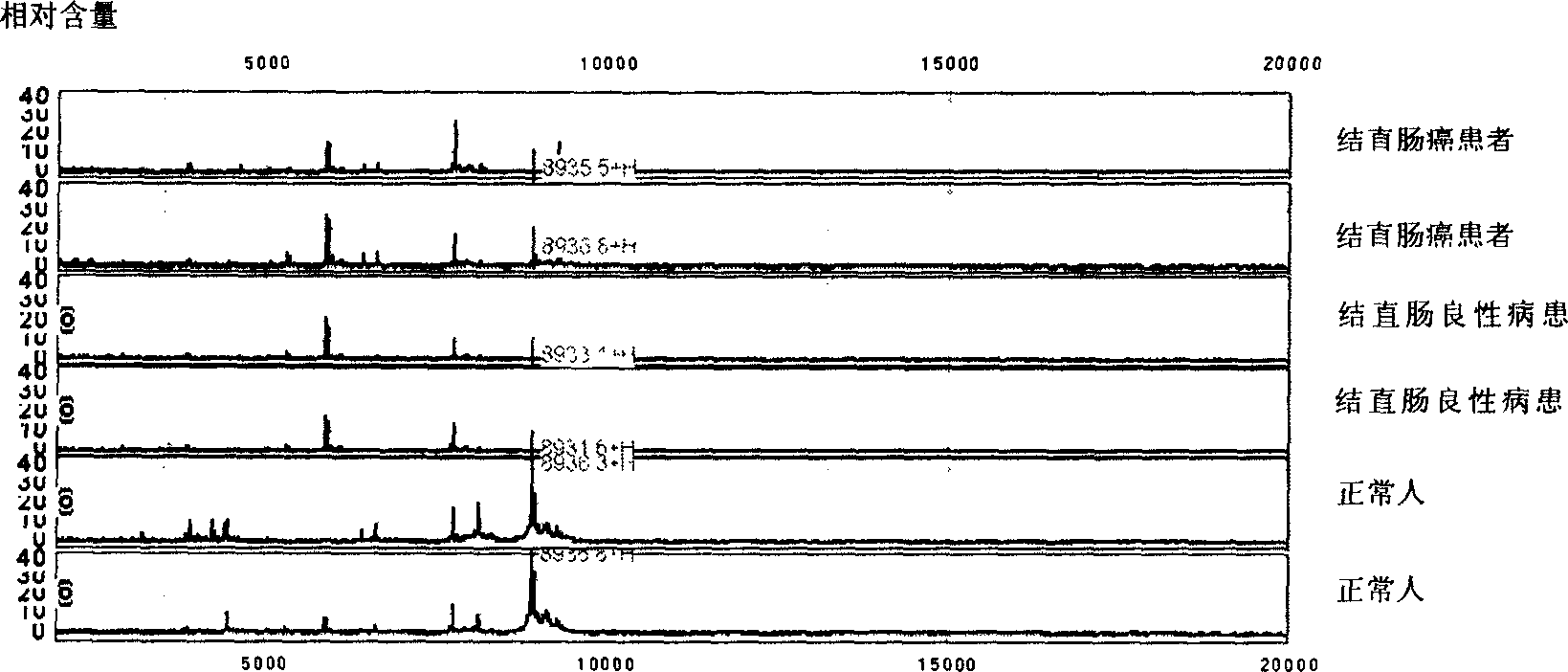
[0063] Example 1 Distinguishing protein fingerprints in blood from normal human cells, cells from patients with colorectal cancer and cells from patients with colorectal polyposis
[0064] (1) Experimental method
[0065] 1. Materials
[0066] 1. Source of specimens: In the colon cancer patient group, there were 146 colorectal cancer patients, all of whom were pathologically diagnosed as colorectal cancer, including 20 cases of Dukes A stage, 126 cases of Dukes B, Dukes C, and Dukes D stages in total. There were 62 cases of normal people in the normal group. Benign colorectal disease control group included 32 patients with benign colorectal disease, including 10 cases of colorectal adenoma, 6 cases of familial polyposis, and 16 cases of colorectal non-neoplastic polyposis. All three groups were between 40 and 60 years old and had similar exposure histories. The male to female ratio in the colon cancer patient group was 97 males and 49 females. Benign colorectal disease gro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com