Fast detection method of pine tree wilt disease pathogenic nematode
A pathogenic nematode and detection method technology, which is applied in the field of forest disease detection, can solve the problems of high price and application limitation, and achieve the effect of convenient operation, high accuracy, and favorable promotion and application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
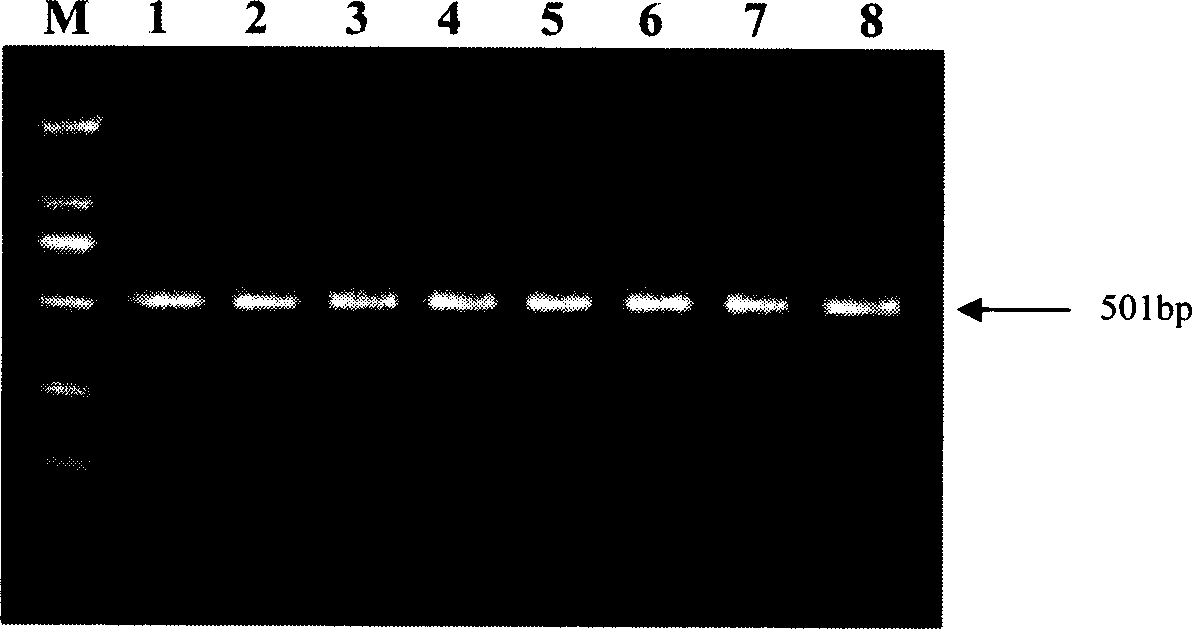
[0020] 1. Design of specific primers for the detection of B. xylophilus and B. xylophilus: 2 populations of B. xylophilus and 5 populations of B. xylophilus from different sources were amplified using the general primers of the nematode ITS, and ITS1, 5.8 containing rDNA were obtained. S, the amplified fragment of ITS2 region; the DNA sequence of each amplified fragment was determined by the dideoxy termination method; the obtained sequence information was analyzed with DNASTAR 5.0 software to find out the difference sequence between the populations of B. xylophilus and B. xylophilus. According to the difference The sequence was designed using primer Premier 5.0 software to design pine xylophilus specific primers BX1: 5'-CGATGATGCGATTGGTGACTT-3' and BX25'-ATGCGAGACGACTGTCACAACG-3', and pine xylophilus specific primers BM1: 5'-TTCTCAAGTTTCTGCATTTGTAAG-3' and BM25' -GGCGCGGTCGCACAATCGAG-3', where BX1 is located in the ITS1 region of rDNA, BX2 is located in the ITS2 region of rDNA...
Embodiment 2
[0025]1. Design of specific primers for the detection of B. xylophilus and B. xylophilus: 2 populations of B. xylophilus and 5 populations of B. xylophilus from different sources were amplified using the general primers of the nematode ITS, and ITS1, 5.8 containing rDNA were obtained. S, the amplified fragment of ITS2 region; the DNA sequence of each amplified fragment was determined by the dideoxy termination method; the obtained sequence information was analyzed with DNASTAR 5.0 software to find out the difference sequence between the populations of B. xylophilus and B. xylophilus. According to the difference The sequence was designed using primer Premier 5.0 software to design pine xylophilus specific primers BX1: 5'-CGATGATGCGATTGGTGACTT-3' and BX25'-ATGCGAGACGACTGTCACAACG-3', and pine xylophilus specific primers BM1: 5'-TTCTCAAGTTTCTGCATTTGTAAG-3' and BM25' -GGCGCGGTCGCACAATCGAG-3', where BX1 is located in the ITS1 region of rDNA, BX2 is located in the ITS2 region of rDNA,...
Embodiment 3
[0030] 1. Design of specific primers for the detection of B. xylophilus and B. xylophilus: 2 populations of B. xylophilus and 5 populations of B. xylophilus from different sources were amplified using the general primers of the nematode ITS, and ITS1, 5.8 containing rDNA were obtained. S, the amplified fragment of ITS2 region; the DNA sequence of each amplified fragment was determined by the dideoxy termination method; the obtained sequence information was analyzed with DNASTAR 5.0 software to find out the difference sequence between the populations of B. xylophilus and B. xylophilus. According to the difference The sequence was designed using the primer Premier 5.0 software to design the specific primers BX1 of B. xylophilus: 5'-CGATGATGCGATTGGTGACTT-3' and BX25'-ATGCGAGACGACTGTCACAACG-3', and the specific primers BM1 of B. xylophilus: 5'-TTCTCAAGTTTCTGCATTTGTAAG-3' and BM2: 5'-GGCGCGGTCGCACAATCGAG-3', in which BX1 is located in the ITS1 region of rDNA, BX2 is located in the I...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com