Method of synchronous distinguishing newcastle disease virus and vaccine virus and identifying virulence and genotype
A newcastle disease virus and vaccine virus technology, applied in the field of automated analysis software, can solve the problems of small number of verification samples, insufficient theoretical foundation, and difficult application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
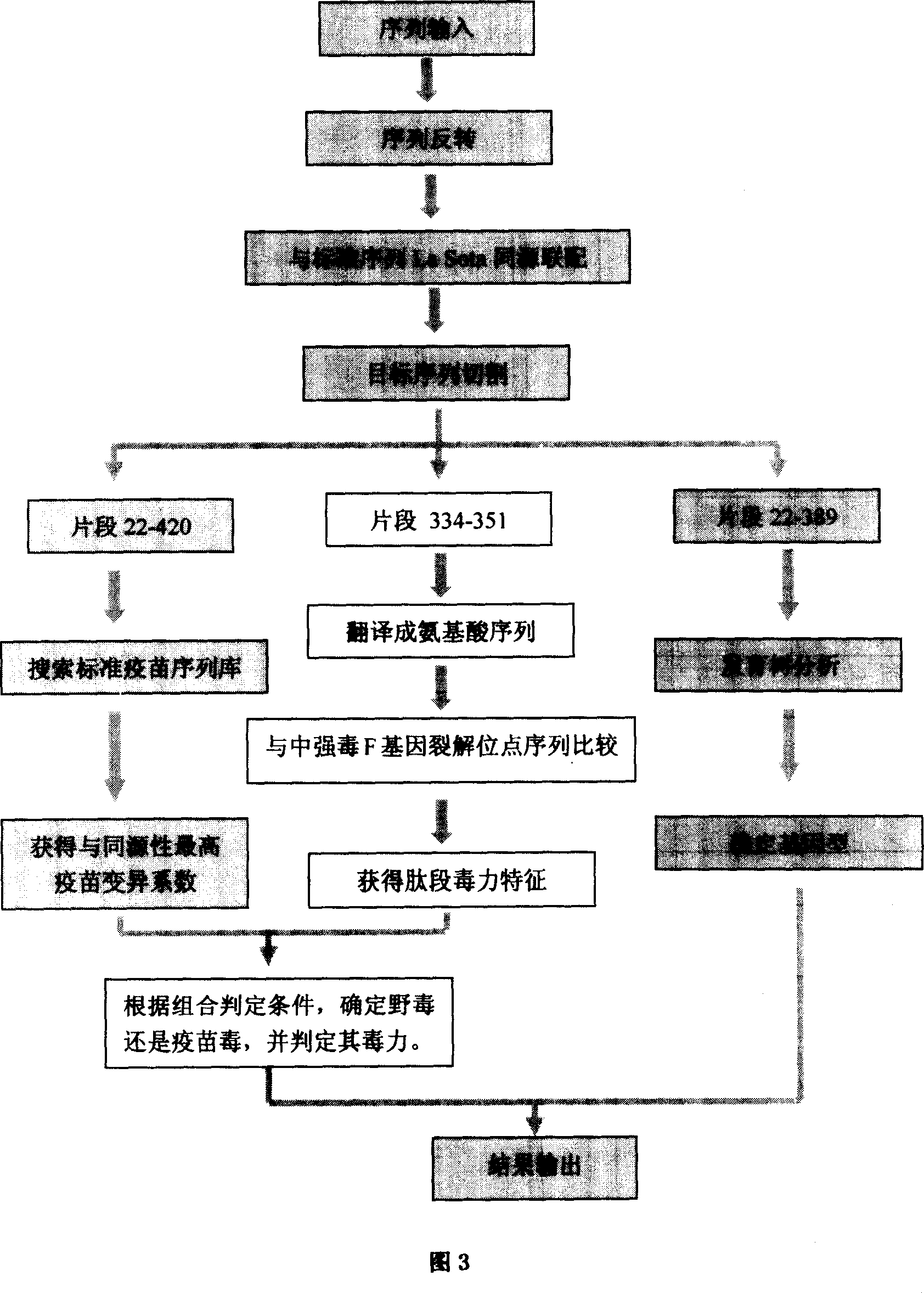
Image
Examples
Embodiment 1
[0116] Embodiment 1-isolate the detection of Newcastle disease strain
[0117] 1. Virus isolation and identification.
[0118] According to the method of "Diagnostic Test and Vaccine Standard Manual" (OIE, 1996), 51 strains of Newcastle disease virus were isolated from geese, pigeons, chickens and ostriches.
[0119] 2. Extraction of viral nucleic acid RNA
[0120] (1) Take 100 μl of allantoic fluid to be tested.
[0121] (2) Add 900 μl TRIzol reagent (Gibco-BRL company), mix well, and keep at room temperature for 5 minutes;
[0122] (3) Add 0.2ml or appropriate amount of chloroform, vortex and mix well, then let stand at room temperature for 3 minutes;
[0123] (4) 12000g, 4°C, centrifuge for 15 minutes;
[0124] (5) Carefully draw the supernatant into a new 1.5ml centrifuge tube;
[0125] (6) Add 0.5ml or an appropriate amount of isopropanol, vortex and mix well, then place at room temperature for 10 minutes;
[0126] (7) 12000g, 4°C, centrifuge for 10 minutes;
[012...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com