Method of detecting cardiac hypertrophy through probe hybridization and gene expression analysis
a probe hybridization and gene expression technology, applied in the field of detecting cardiac hypertrophy through probe hybridization and gene expression analysis, can solve the problems of relatively little information available, myocardial hypertrophy, and impair myocardial stiffness, and achieve the effects of increasing ventricle mass, treating and preventing cardiac hypertrophy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
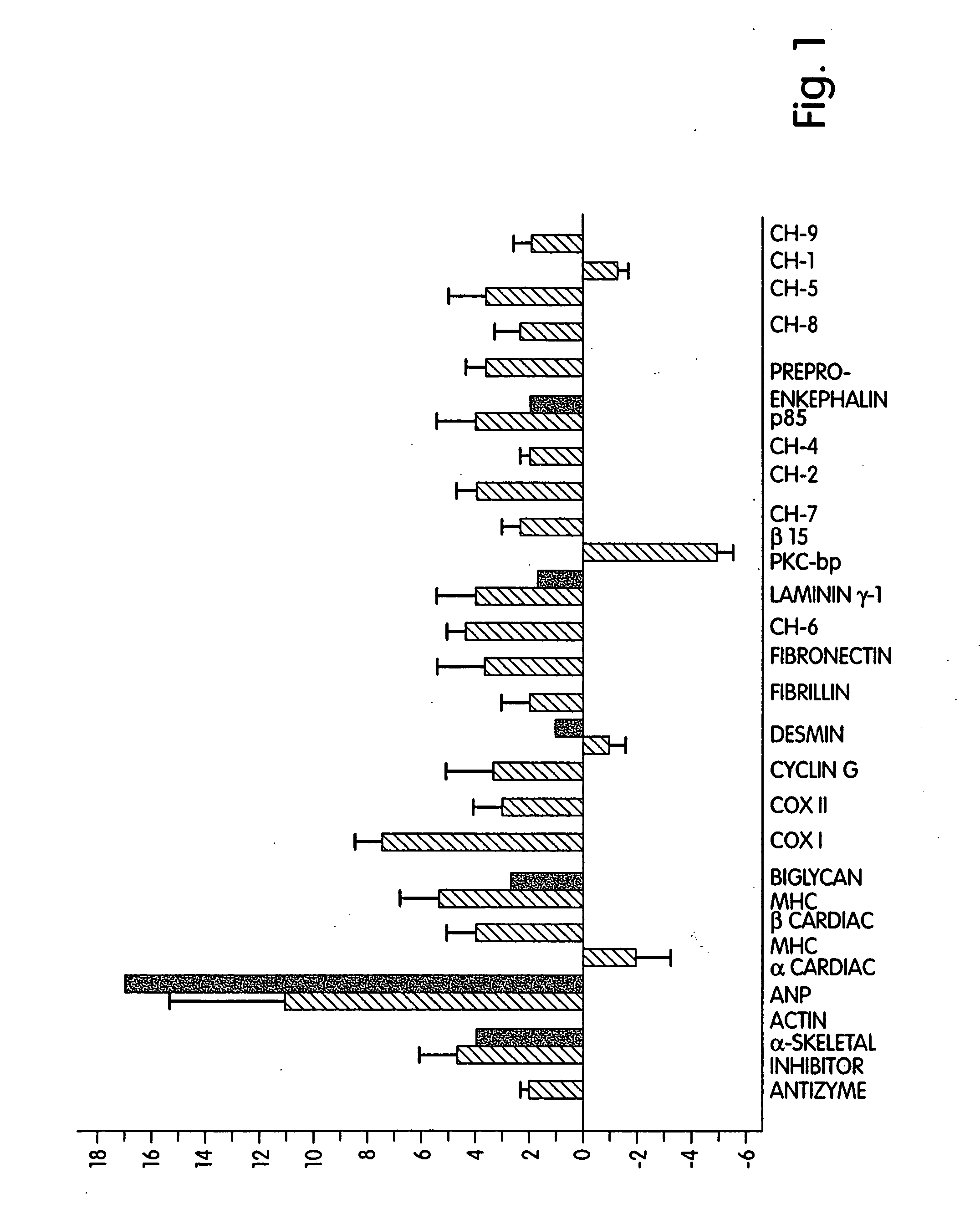
[0195] To obtain a more detailed understanding of the changes in gene expression occurring during pressure overload hypertrophy, QEA was used to identify expression differences in a rat surgical model of pressure overload (POL) induced cardiac hypertrophy. This in vivo model is attractive for expression analysis as there are limited changes in tissue cellularity by infiltration of non-resident cells. The tissue reaction to acute POL is primarily due to an intrinsic response of the myocardium including hypertrophy of the cardiomyocytes, in addition to hyperplasia of endothelial, smooth muscle, and mesenchymal cells (Weber and Brilla, Circulation 83:1849-65 (1991); Cooper, IV, Ann. Rev. Physiol. 49:501-18 (1987)). While this process is initially adaptive, there is ultimately a deterioration of contractile function accompanied by interstitial and perivascular fibrosis and increased wall stiffness (Kimura et al., Heart Circ. Physiol. 25:H1006-H1011 (1989); Batra and Rakusan, J. Cardiova...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| body weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com