Detection of methylated CpG rich sequences diagnostic for malignant cells
a methylated cpg and rich sequence technology, applied in the direction of fluid pressure measurement, liquid/fluent solid measurement, peptide measurement, etc., can solve the problem of still uncertain methylation status of multiple cpg islands
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
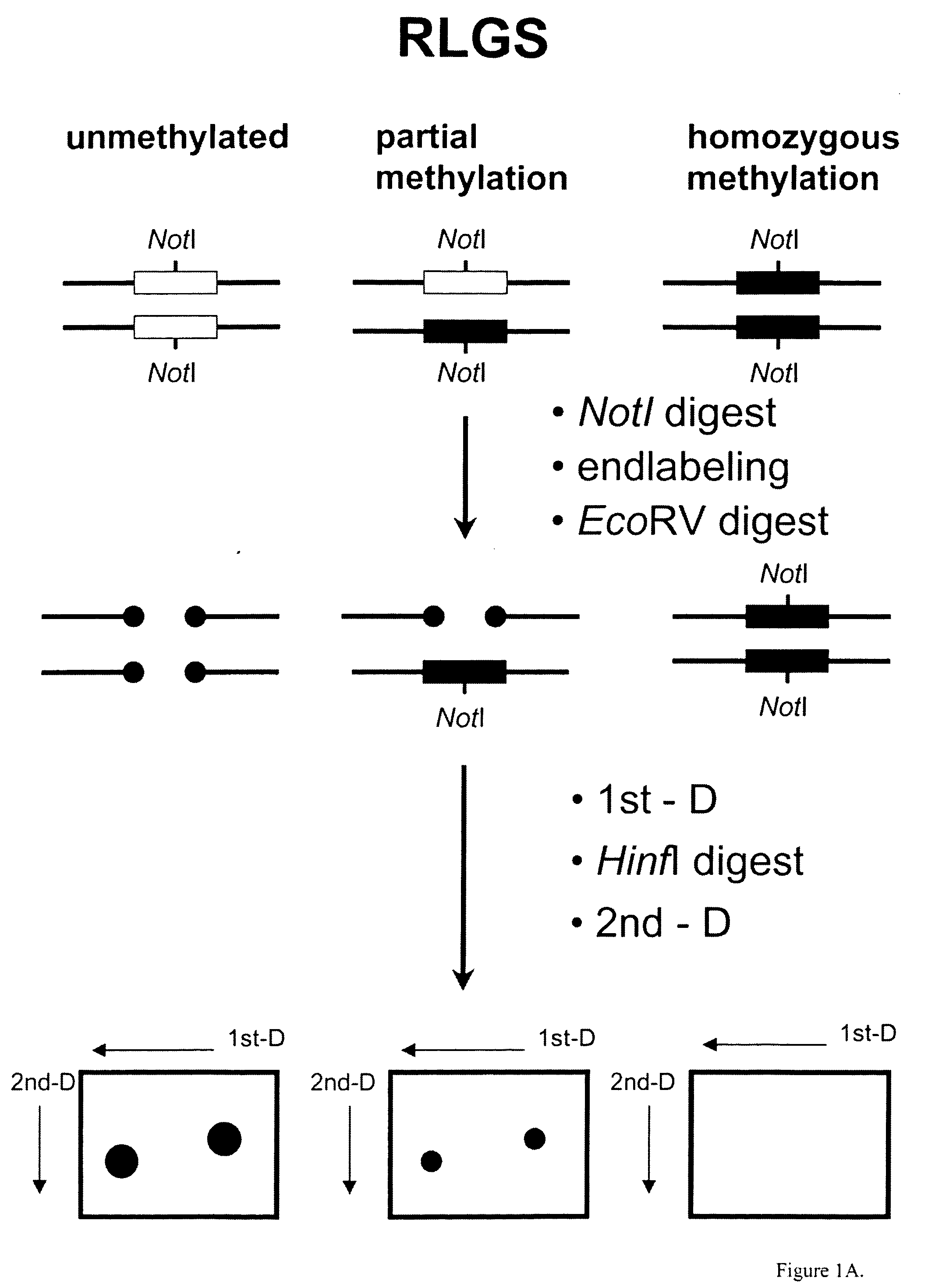
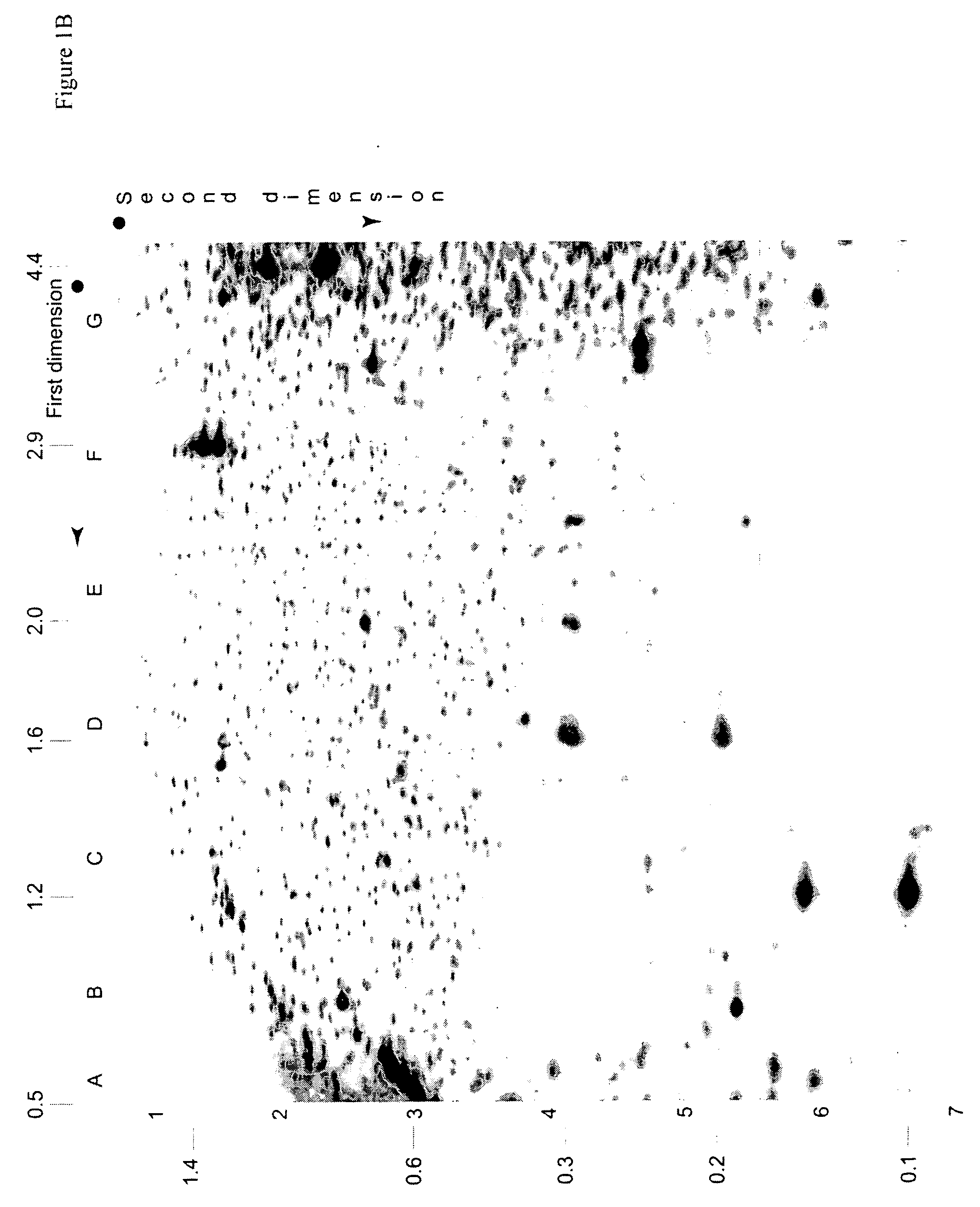
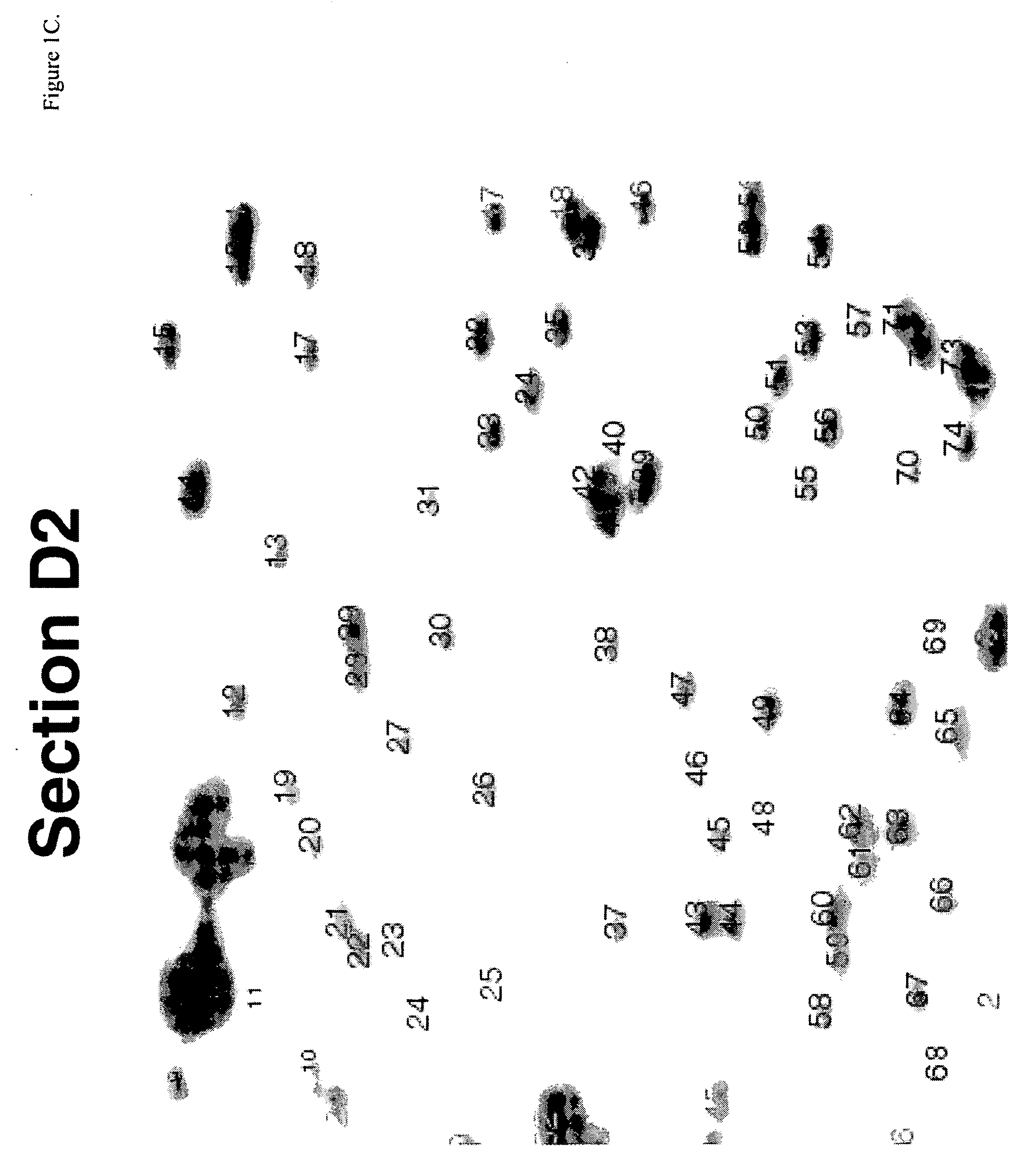
Identification of Diagnostic Markers Using NotI and RLGS
A. Isolation and Enzymatic Processing of Genomic DNA
[0053] Tissue from solid tumors was obtained as surgical tissue samples. Where possible, surrounding non-tumor tissue was taken and used as a control. Where it was not possible to obtain patient-matched normal tissue, normal tissue from multiple patients was used. Tissue samples from patients with acute myelogenous leukemis (AML) consisted of either bone marrow aspirates or peripheral blood. Normal samples were obtained from the same patients who were in remission after chemotherapy.
[0054] The surgically removed tissues were quickly frozen in liquid nitrogen and stored at −80° C. prior to isolation of DNA. When DNA was ready to be isolated, 2 ml of lysis buffer (10 mM Tris, pH 8.0; 150 mM EDTA, 1% sarkosyl) was added to 100-300 mg of tissue in a 50 ml Falcon tube and frozen in liquid nitrogen. The frozen mixture was then removed from the tube, wrapped in aluminum foil, and...
example 2
Identification of Diagnostic Markers for Lung Cancer Using AscI and RLGS
[0088] Tissue from lung tumors was obtained as surgical tissue samples. Where possible, surrounding non-tumor tissue from the same patient was obtained and used as a control. DNA was isolated from the tissue as described in Example 1. In preparation for RLGS analysis, the ends of the DNA were blocked as described in Example 1. The DNA was then digested with AscI followed by digestion with EcoRV. The AscI restriction enzyme recognizes the sequence 5′GGCGCGCC3′ and does not cleave said sequence if cytosines within the sequence are methylated. First dimension gel electrophoresis, in-gel digestion with HinfI, second dimension gel electrophoresis and autoradiography were performed as described in Example 1.
[0089] RLGS profiles from lung tumor DNA were compared with RLGS profiles obtained from healthy, non-tumor tissue DNA. Spots which were lost or present at reduced intensity in tumor tissue RLGS profiles as compar...
example 3
Design of Primers for Cancer Diagnosis
[0091] Primers are designed for diagnosis of cancer using methylation-specific PCR (MSR). The primers are designed to amplify regions of the human genome whose sequences are contained within the library clones disclosed in this application. Two sets of primers are needed for each library clone whose DNA sequence is to be used for diagnosis of cancer. Each primer set is designed to amplify the same region of the genome, said region beginning at the end of a library clone containing the methylation-sensitive restriction enzyme recognition site (i.e., the NotI site for the library described in Example 1; the AscI site for the library described in Example 2) and ending at a region contained within the clone up to 200 nucleotides from the methylation-sensitive restriction enzyme recognition site.
[0092] The first set of primers is designed to amplify template genome DNA whose cytosine residues are not methylated and, after bisulfite treatment, the c...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com