Methods and compositions for preparing capped RNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Standard mMmM In Vitro Transcription Reaction
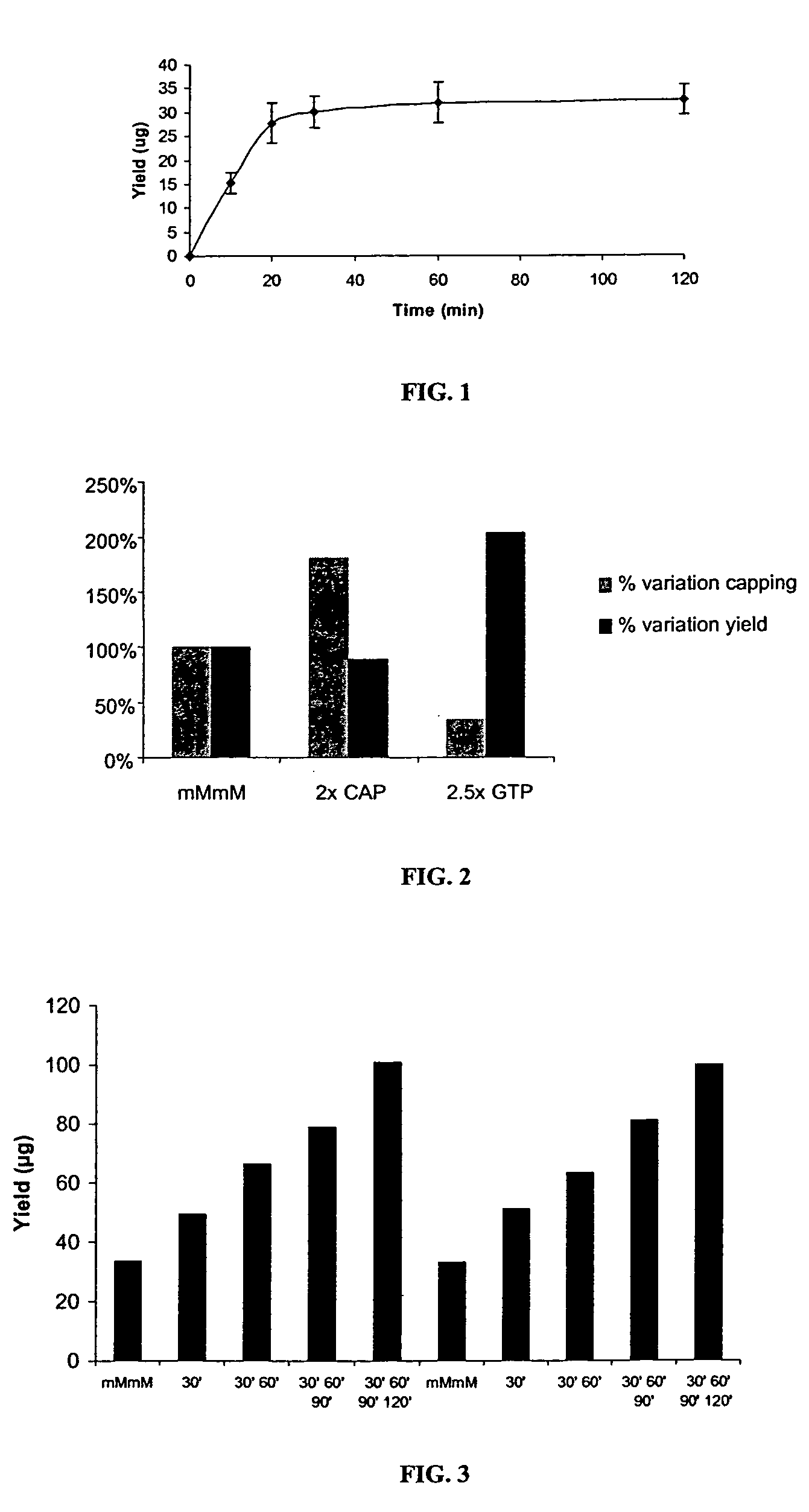
[0104] A standard mMESSAGE mMACHINE® T7 Kit reaction contains 50 ng / μl plasmid template; 4 U / μl T7 RNA polymerase; 0.005 U / μl IPP; 0.03 U / μl RNase Inhibitor; 0.01 U / μl SUPERase•In; 0.1% Chaps; 40 mM Tris, pH 8.0; 20-30 mM MgCl2; 2 mM spermidine; 10 mM DTT; 7.5 mM ATP; 7.5 mM CTP; 7.5 mM UTP; 1.5 mM GTP; and, 6 mM m7GpppG cap analog. Components are assembled at room temperature in a final volume of 20 μl and the reaction is incubated at 37° C. up to two hours. Under these conditions, transcription reactions with pTRI-Xef (˜1.8 kb RNA), pAmbluc (˜1.8 kb RNA) or p4kb (˜4 kb RNA) templates produce ˜30 μg of transcript (˜1.5 mg / ml) in 30 minutes. A time course study with the p4kb template is presented in FIG. 1. Analysis of the RNA produced on a RNA LabChip with the Agilent 2100 bioanalyzer showed no RNA degradation over time.
[0105] The cap:GTP ratio in a standard mMmM reaction is 4:1. Analysis of the capping efficiency confirmed that less t...
example 2
Fed-Batch In Vitro Transcription Reaction—Manual Feed
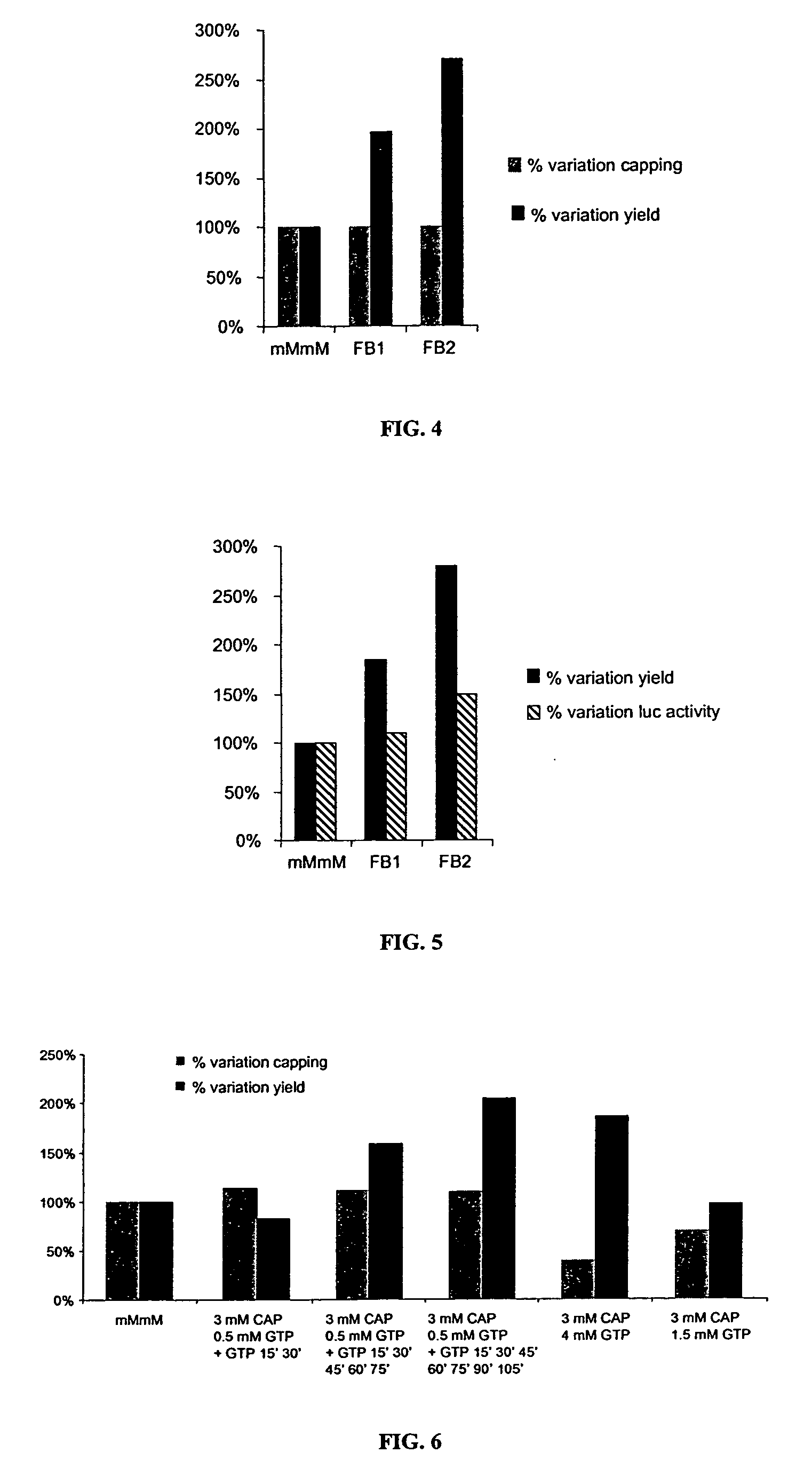
[0107] The time course study presented in FIG. 1 shows that a standard mMmM reaction is essentially completed after 30 min incubation at 37° C. At this time point, most of the GTP has been incorporated into the transcript. In contrast, only a small fraction of the cap analog has been used as 1) there is a 4-fold excess of cap over GTP in the reaction, 2) only 1 molecule of cap is incorporated per molecule of RNA synthesized, and 3) only ˜80% of the transcripts are capped. The amount of GTP used and the percentage of cap used in the reaction can be easily estimated from the yield and the size of the transcript. The average molecular weight of a given RNA molecule is equivalent to its total number of residues. If this value is multiplied by 320 g / mol (the average molecular weight for all 4 residues), then:
[GTP] used in mM=(3.12×yield×G) / nt
% cap used=(250×yield) / (nt×[cap]) [0108] with yield in μg / μl or mg / ml [0109] G=number of G r...
example 3
Fed-Batch In Vitro Transcription Reaction—Other Cap Analogs
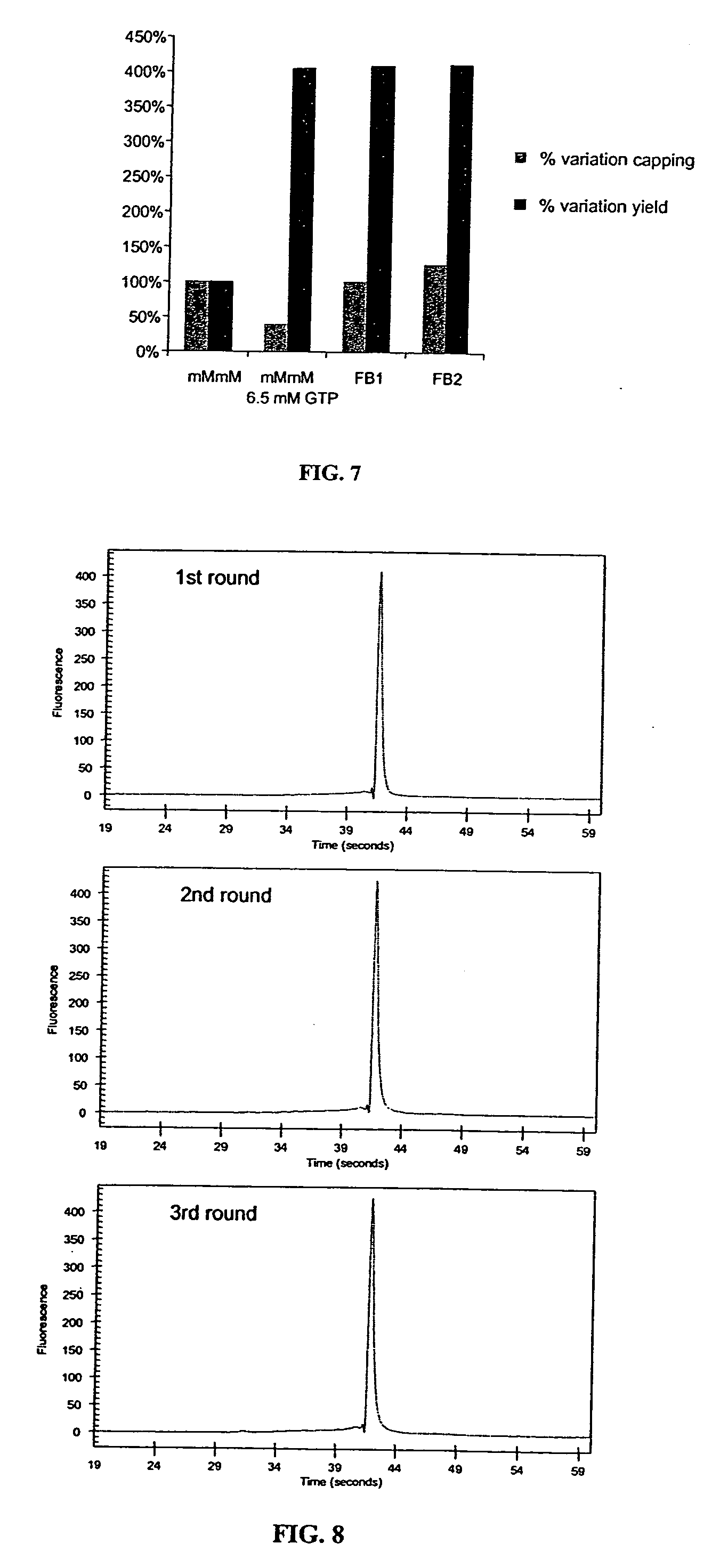
[0114] Any non-extending, mono- or di-nucleotide (i.e., that cannot be incorporated as a 3′ nucleotide in a transcription reaction) can be incorporated as the first nucleotide of a transcript by phage RNA polymerases, and are compatible with the fed-batch strategy. This includes 5′ hydroxyl, monophospate or biotinylated nucleotides, trimethylated cap analog (m2,2,7GpppG), unmethylated cap variant (GpppG), tetraphosphate cap variant (m7GppppG) or other cap variants (e.g. m7GpppA, m7GpppC). Of particular interest are the anti-reverse cap analogs (ARCAs). With the standard cap analog m7GpppG, because of the presence of a 3′-OH on both the m7Guanosine and Guanosine moieties, 30-50% of the initiating dinucleotide is incorporated in a reverse, non-functional orientation (Pasqinelli et al., 1995). ARCA molecules such as m7dGpppG, m7,3′-OMeGpppG, m7,3′-OMeGppppG or m7,2′-OMeGppppG (Stepinski et al., 2001; Jemielity et al., 2003) ar...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molar density | aaaaa | aaaaa |
| Molar density | aaaaa | aaaaa |
| Molar density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com