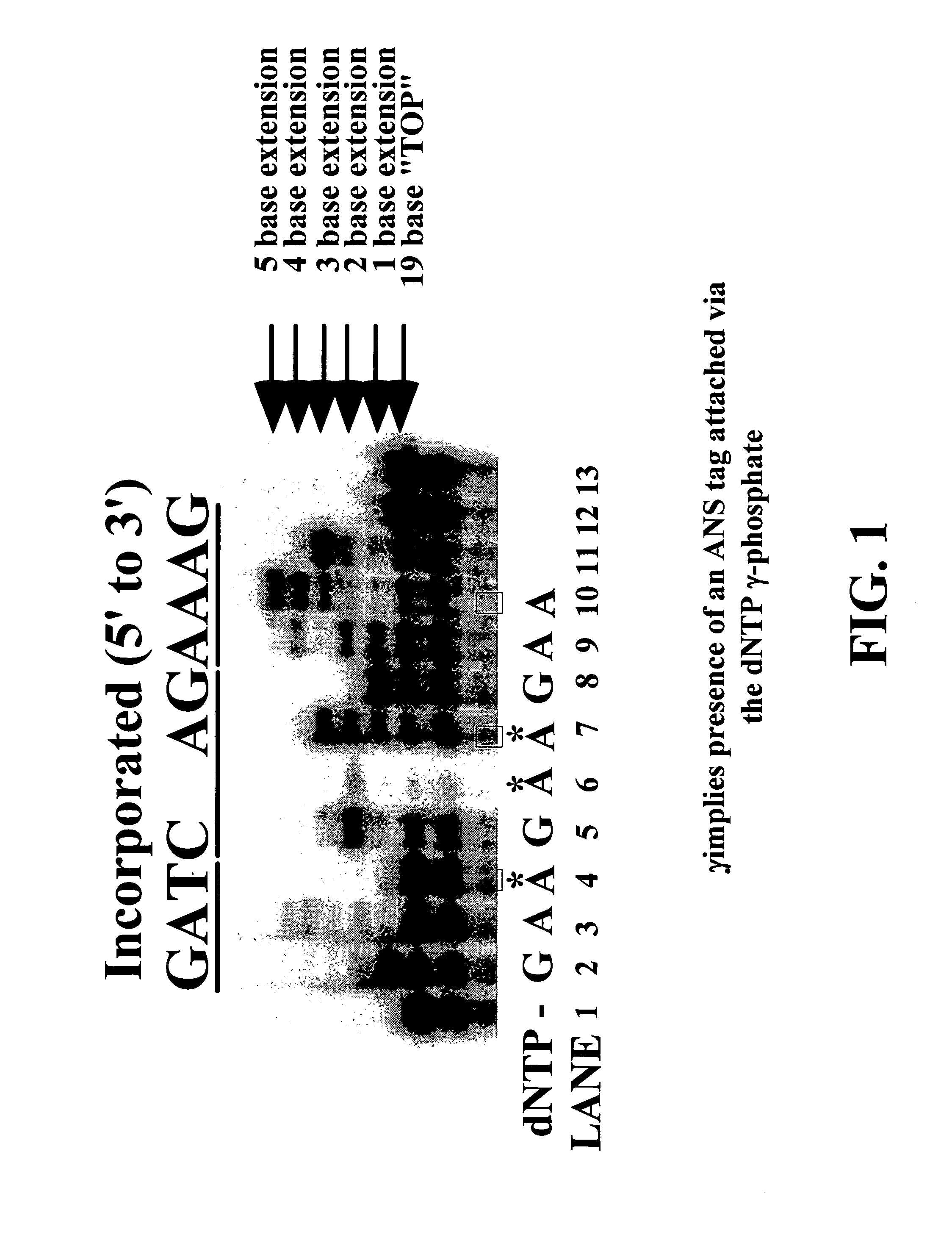
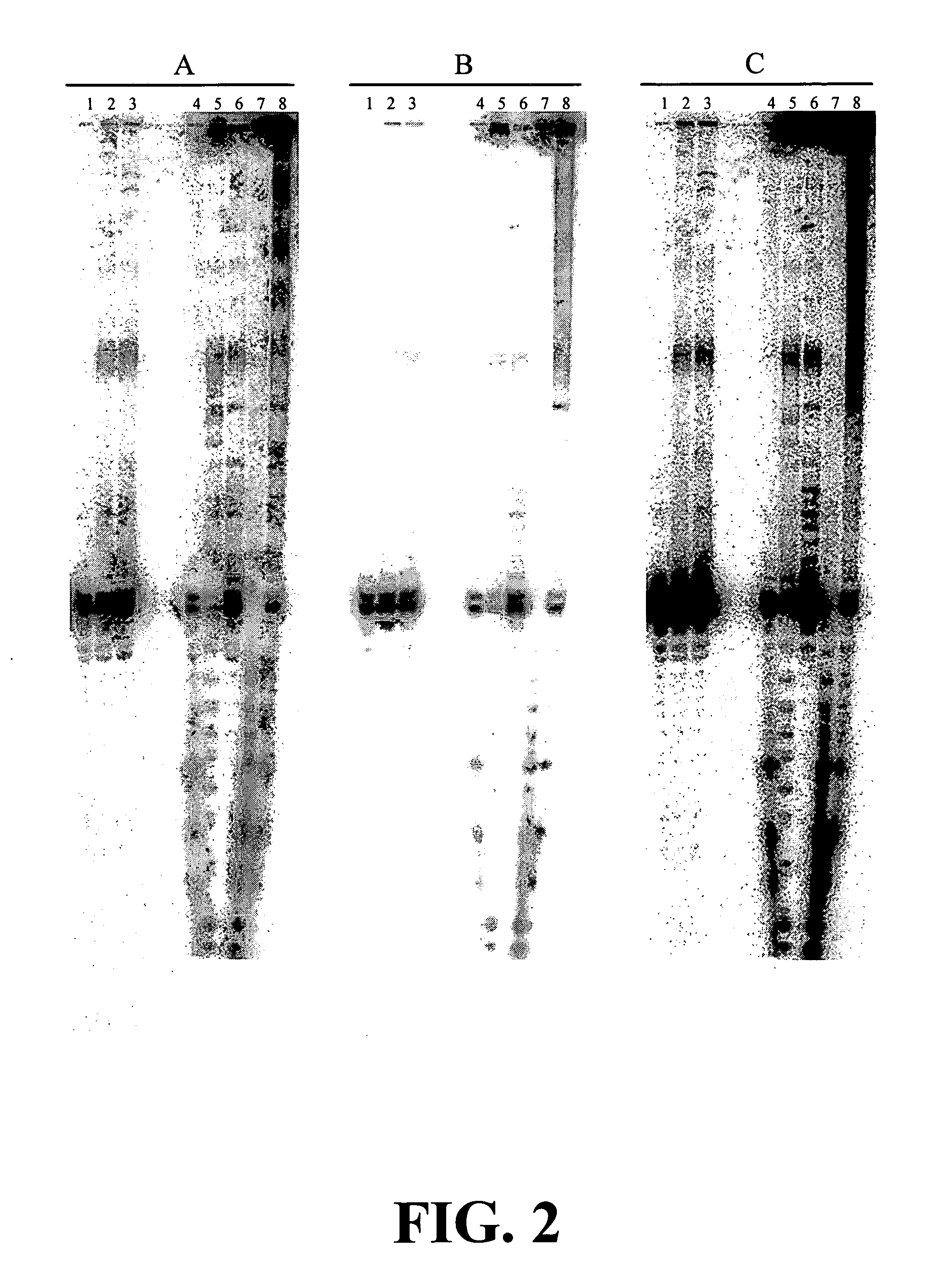Enzymatic nucleic acid synthesis: methods for inhibiting pyrophosphorolysis during sequencing synthesis
a technology of pyrophosphorolysis and sequencing synthesis, which is applied in the field of enzyme-based nucleic acid synthesis, can solve the problems of pyrophosphorolysis, where an oligonucleotide is reduced in length, and the fluorescent efficiency of the halogen quencher used in the assay is very low, so as to enhance the progress of the reaction, prevent pyrophosphorolysis, and improve the effect of the reaction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0082] The inventors have found that nucleotide monomers or analogs thereof bearing an atomic and / or molecular tag on a site of the molecule can increase the fidelity of nucleotide polymerization for nucleotide polymerization agents that can incorporated the modified monomers. This increase in fidelity is useful for improving nucleic acid sequencing determinations using any of the standard sequencing reactions such as PCR, rolling circle or the like. Additionally, these modified monomers may allows the construction of drugs for animal or human use that would increase the fidelity of viral disease replication in vivo decreasing mutagenesis allowing the immune system to recognize the virus. Such a medication may be of particular benefit for virus such as the HIV virus that causes AIDS.
[0083] Mutation of amino acids within the polymerase is the classic approach to understand enzyme action and / or modulate enzyme fidelity (Yang S and Chatterjee D K. (1999) PCT WO9910366; Wainberg M A, D...
PUM
| Property | Measurement | Unit |
|---|---|---|
| contour length | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com