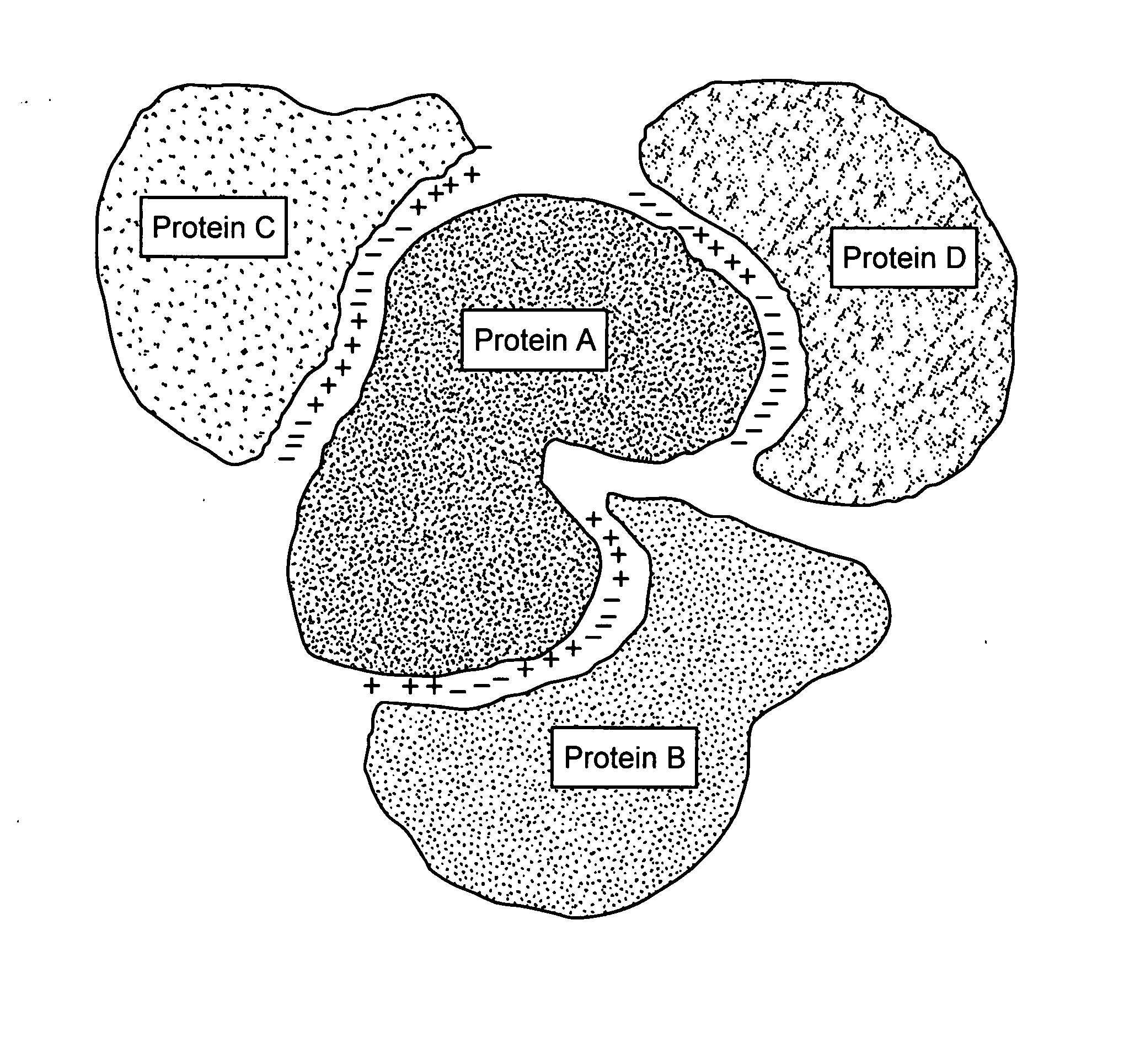
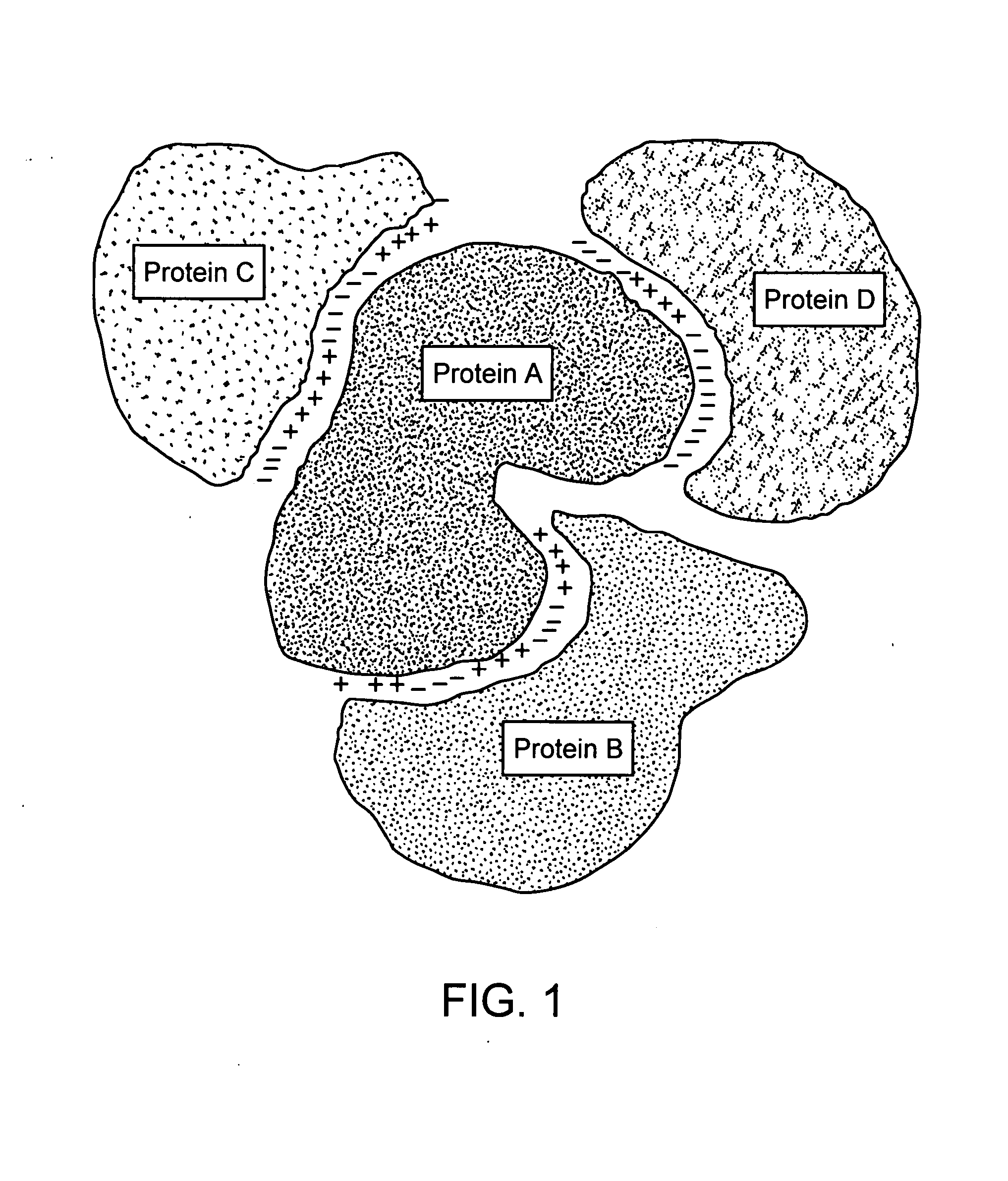
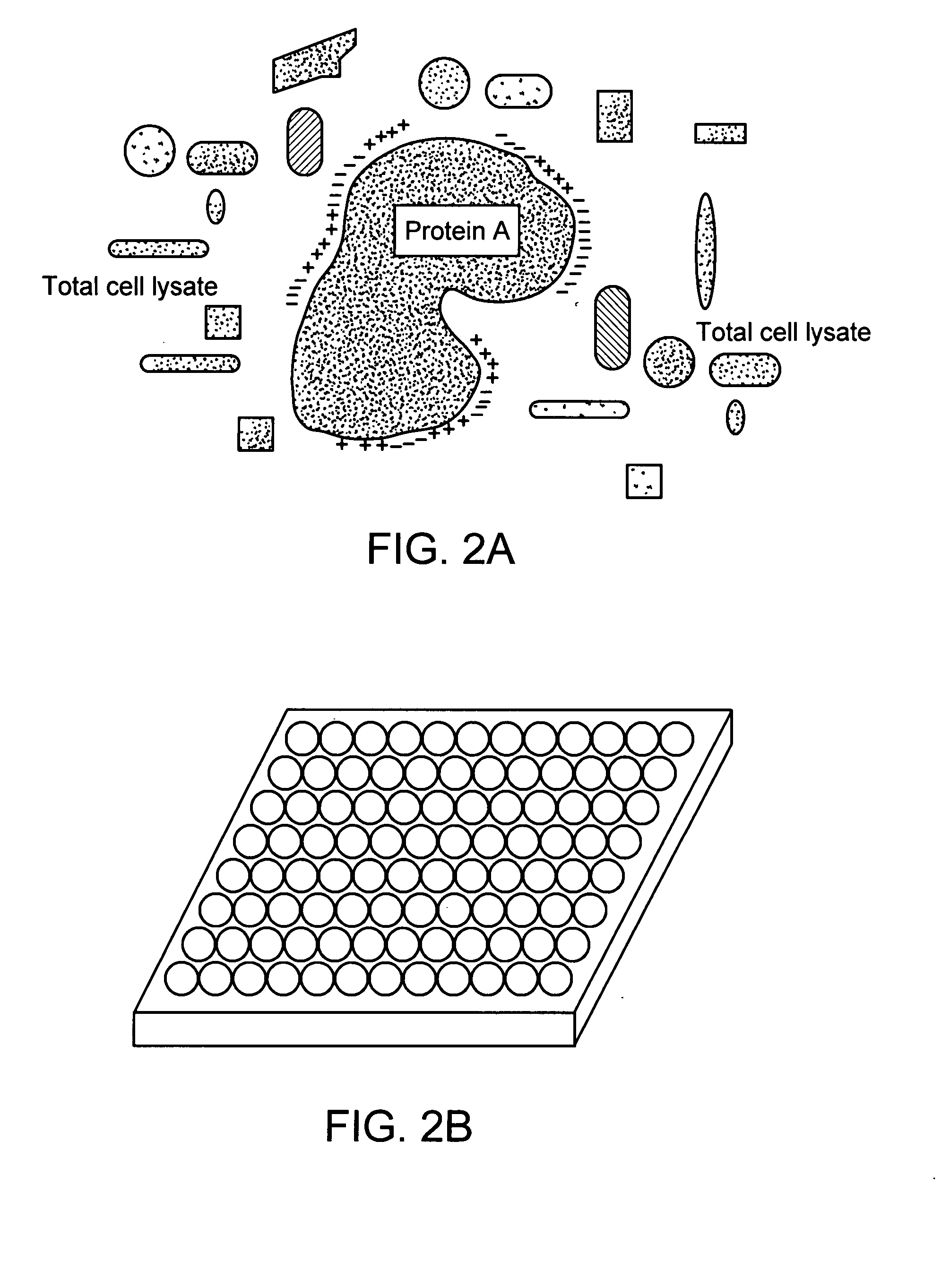
Protein-protein interactions and methods for identifying interacting proteins and the amino acid sequence at the site of interaction
a technology of protein-protein interactions and amino acid sequences, applied in the field of proteomics, can solve the problems of most research and development, target protein identification and functional site identification, and development of treatmen
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
P-glycoprotein Binding to Tubulin is Mediated by Sequences in the Linker Domain
[0110] The successful treatment of cancer patients with chemotherapeutic drugs is often limited by the development of drug-resistant tumors. Tumor cell lines selected, in vitro, with a single anticancer drug become resistant to a broad spectrum of chemotherapeutic drugs, termed multidrug resistant (or MDR) tumor cells (for review, (21, 45, 66). Moreover, the expression of MDR in these tumor cells has been associated with the overexpression of two membrane proteins; the MDR1 P-glycoprotein (P-gp) and the multidrug resistance-associated protein (MRP1) (21, 45, 66). Both P-gp and MRP are members of a large family of membrane transporter proteins known as ATP binding cassette proteins or ABC membrane transporters (54). Although, the structure of P-gp1 remains a matter speculation (91), cumulative topological evidence suggest a tandemly duplicated structure of six transmembrane domains and a large cytoplasmic...
example 2
Materials
[0116] [35S] methionine (1000 Ci / mmol; Amersham Life Sciences, Inc.) and [125] goat anti-mouse antibody were purchased from Amersham Biochemical Inc. Protein-A Sepharose-4B was purchased from Bio-Rad Life Science. All other chemicals used were of the highest commercial grade available.
example 3
Peptide Synthesis
[0117] Pre-derivatized plastic rods, active ester and polypropylene trays were purchased from Cambridge Research Biochemicals (Valley Stream, N.Y.). Peptides were synthesized on solid polypropylene rods as previously described (36, 37). Briefly, the F-moc protecting group on the prederivatized polypropylene rods as solid support (arranged in a 96-well formate) was removed by incubation with 20% (v / v) piperidine in dimethylformamide (DMF) for 30 minutes with shaking. Following the deprotection of the β-alanine spacer on the polypropylene rods, Fmoc protected amino acids were dissolved in HOBt / DMF and added to the appropriate wells containing deprotected rods. Coupling of amino acids was allowed to take place for 18 hours at room temperature after which the rods were washed in DMF (1×2 minutes), methanol (4×2 minutes), and DMF (1×2 minutes). The coupling of the second amino acid required the deprotection of the F-moc amino protecting group of the first amino acid and...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| /v/v | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com