Yeast arrays, methods of making such arrays, and methods of analyzing such arrays
a technology of arrays and yeast, applied in the field of yeast arrays and high density output arrays, can solve the problems of phenotypic analysis of the set of viable deletion strains within certain species of yeast, the absence of obvious functions for a large fraction of encoded proteins will quickly become an enormous problem in biology, and the scope of the challenge is immens
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
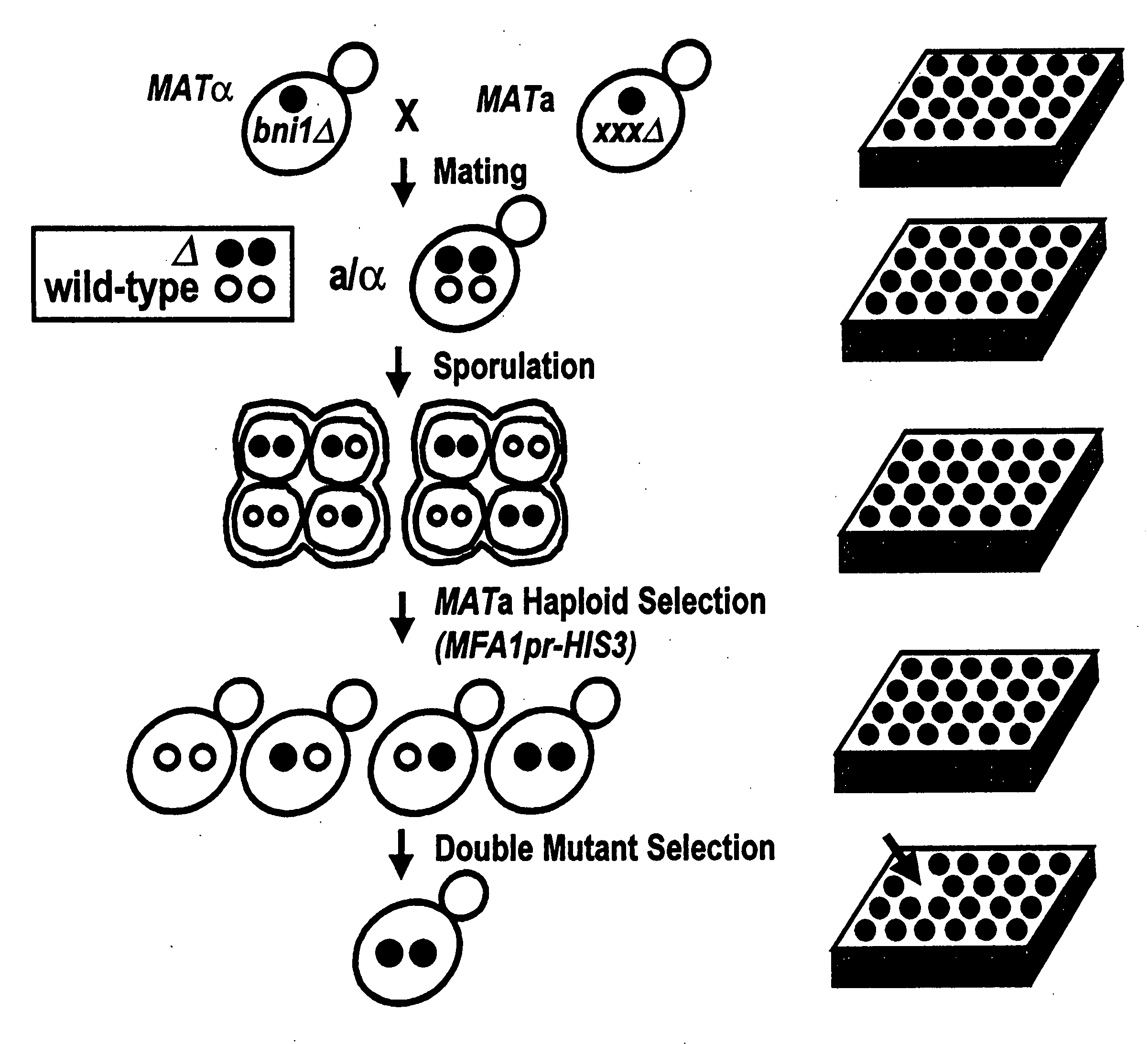
Creation of Double Mutant Output Array
[0086]An example of a starting strain that could be used in this embodiment is the Saccharomyces cerevesiae strain termed Y2454. The Y2454 strain is characterized by being a MAT□ mating type with ura3, leu2, his3, and lys2 mutations, and a HIS3 gene linked to an MFA1 promoter. The ura3, leu2, his3, and lys2 mutations require the strain to be grown in supplemented media to survive. They also carry a can1 null allele which confers canavinine resistance to the cells. A mutant gene, for example, one of the approximately 5,000 non-lethal mutations found in Saccharomyces cerevesiae, is introduced into this strain. The deleted gene is being replaced by a NAT gene which confers noureseothricin resistance to these cells.
[0087]This strain can be crossed with a starting array of yeast strains of the MATa mating type. The strains in this starting array contain ura3, leu2, his3, and met15 knockouts, so that they can only survive on supplemented media. These ...
example 2
Use of Robotics to Generate Output Array
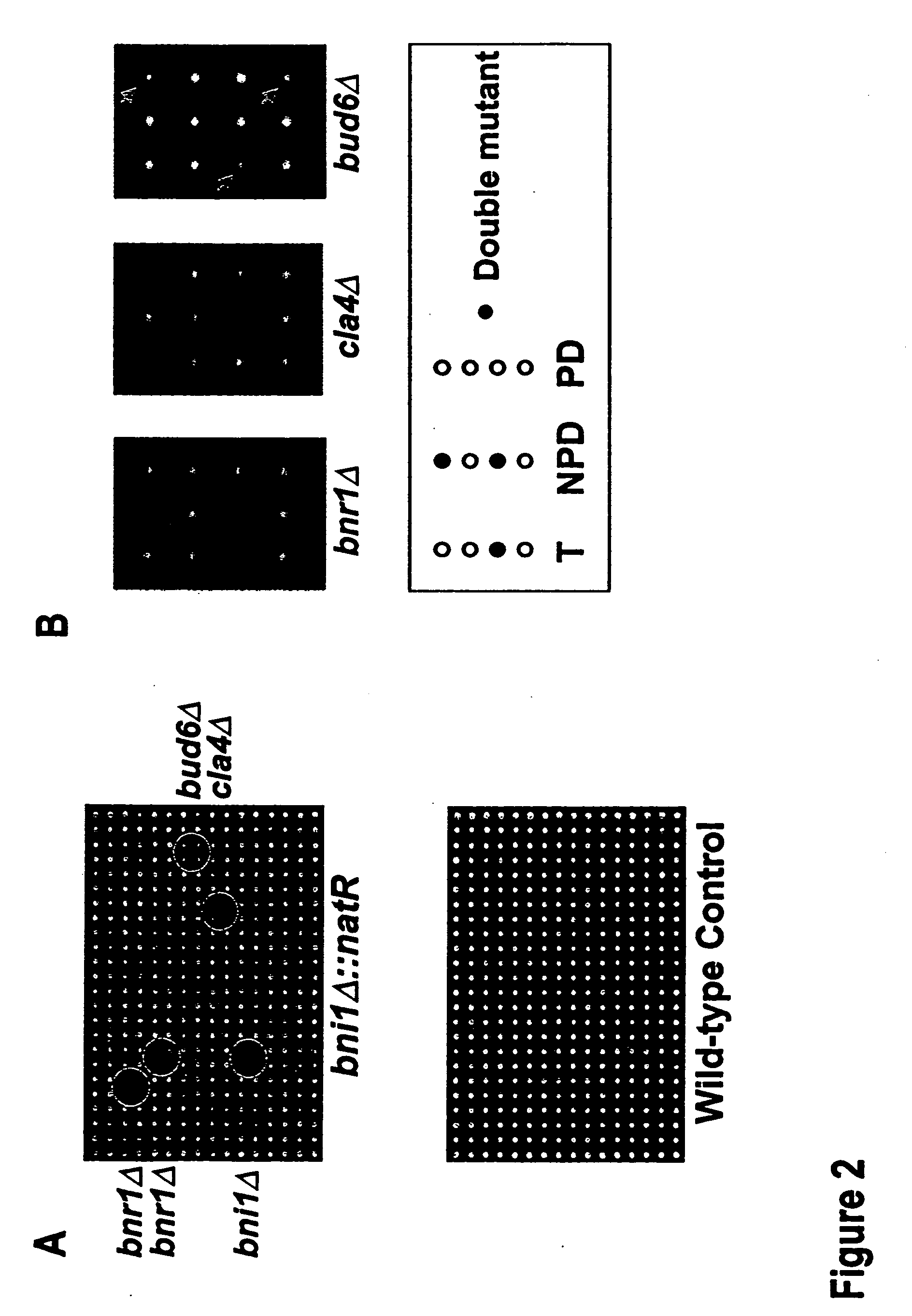
[0096]Following construction of a natR-marked mutant for synthetic lethal analysis, simple replica plating or pinning manipulations will enable us to complete steps 2-6 of Example 1. These steps can be carried out through the use of a robotic colony arrayer, as described below. The Colony Arrayer used in this Example, the Virtek Vision CPCA was designed by our labs in conjunction with Virtek Vision. The CPCA is based upon a system used for genome-wide two-hybrid arrays, since the manipulations required for automated two-hybrid screens are very similar to those required for automated double-mutant construction and synthetic lethal screens.
[0097]For a rapid genome-wide two-hybrid screening procedure, we have created a robotic colony array which includes replica-plate pinhead that transfers 768 individual colonies from one standard microtiter-sized agar-slab plate to another in a single move. This cell density allows efficient mating, on the orde...
example 3
Recovery of Haploid Spore Progeny
[0104]The recovery of haploid spore progeny is mentioned in step 5 in Example 1, and is described in greater detail below. The MATα starting strain described in Example 1, Y2454, carries two selectable markers, can1Δ0 and MFA1pr-HIS3, both of which permit efficient recovery of haploid spore progeny.
[0105]i) MFA1pr-HIS3
[0106]MFA1 encodes the a-factor precursor, which is expressed constitutively in MATa cells. The MFA1 promoter, MFA1pr, is repressed in MATa and MATa / α cells. The MFA1pr-HIS3 reporter was constructed by replacing the MFA1 ORF (open reading frame) with the HIS3 ORF such that MFA1pr drives HIS3 expression. The MATα starting strain, Y2454, fails to grow on synthetic medium lacking histidine because the MFA1pr-HIS3 reporter is repressed in MATα cells. The cells constructed in step 2 of Example 1 will also fail to grow on synthetic medium lacking histidine because the MFA1pr-HIS3 reporter is repressed in MATa / α cells. Following sporulation of...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com