Identification of Compounds Modifying A Cellular Response
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
General Methods for Solid Phase Peptide Synthesis (SPPS)
General for Chemical Synthesis:
[0316]All chemicals described, commercially available and used without further purification. All solvents were HPLC-grade. PEGA-resins were purchased from VersaMatrix A / S, Copenhagen. Each washing step lasted 2 min unless otherwise stated. Purifications were performed on a standard reverse phase HPLC using gradients of acetonitrile-Water with various amounts of TFA.
Coupling of HMBA Linker to PEGA-Resin:
[0317]Dry PEGA-resin was swelled in DCM and washed with DMF (3×). 3.0 eq. HMBA, 2.9 eq. TBTU and 3.0 eq. NEM were mixed in appropriate DMF and allowed to react for 10 min. The mixture was added to resin and after 2 h the resin was washed with DMF (6×), DCM (6×) and lyophilised.
General Procedure for Coupling of Amino Acid to HMBA-Linker:
[0318]Dry PEGA-resin with HMBA-linker was swelled in DCM. 3.0 eq. Fmoc-protected amino acid, 2.25 eq. Melm and 3.0 eq. MSNT were mixed in appropriate amount of DCM an...
example 2a
Screening of Adhesion Peptide Library
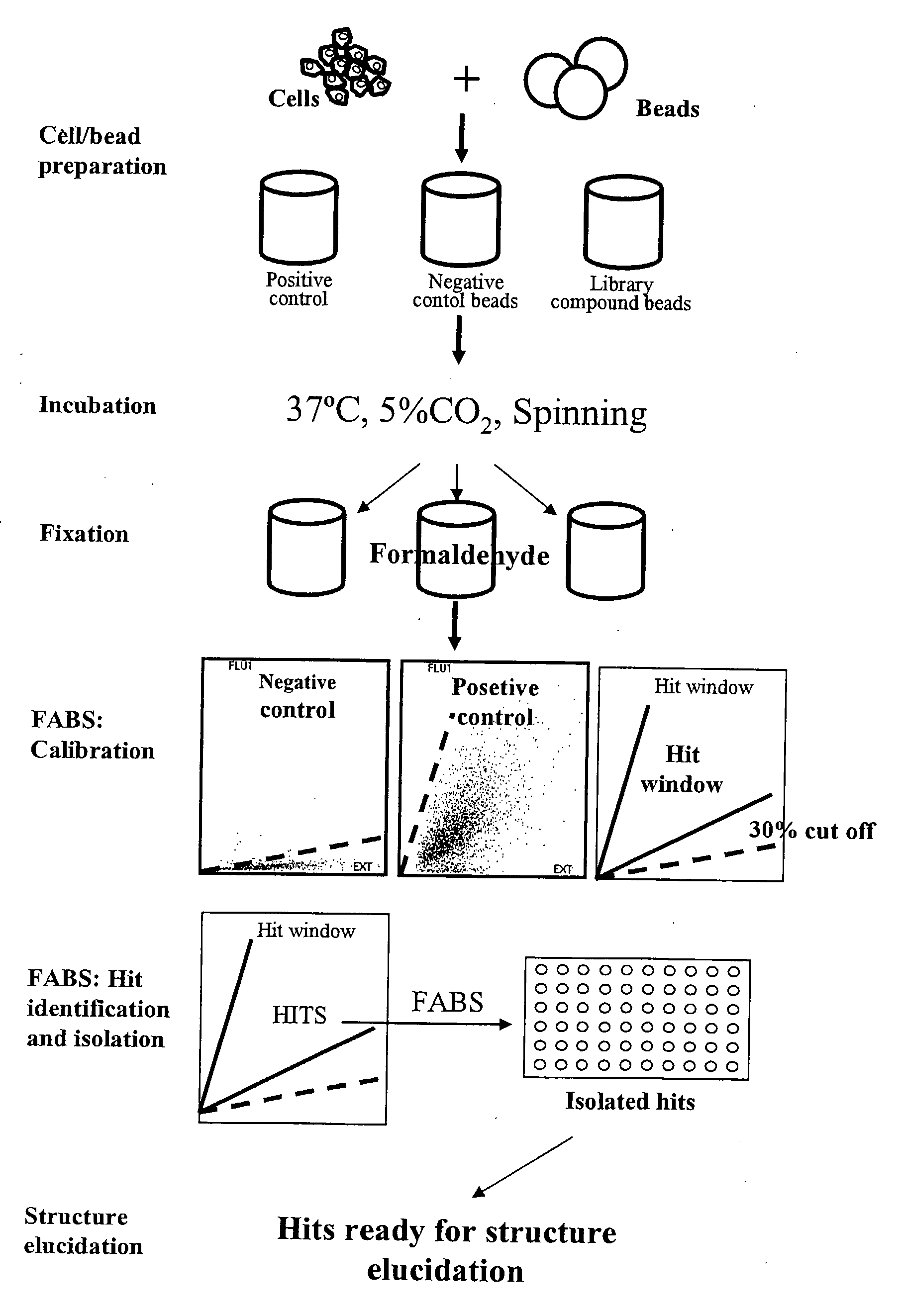
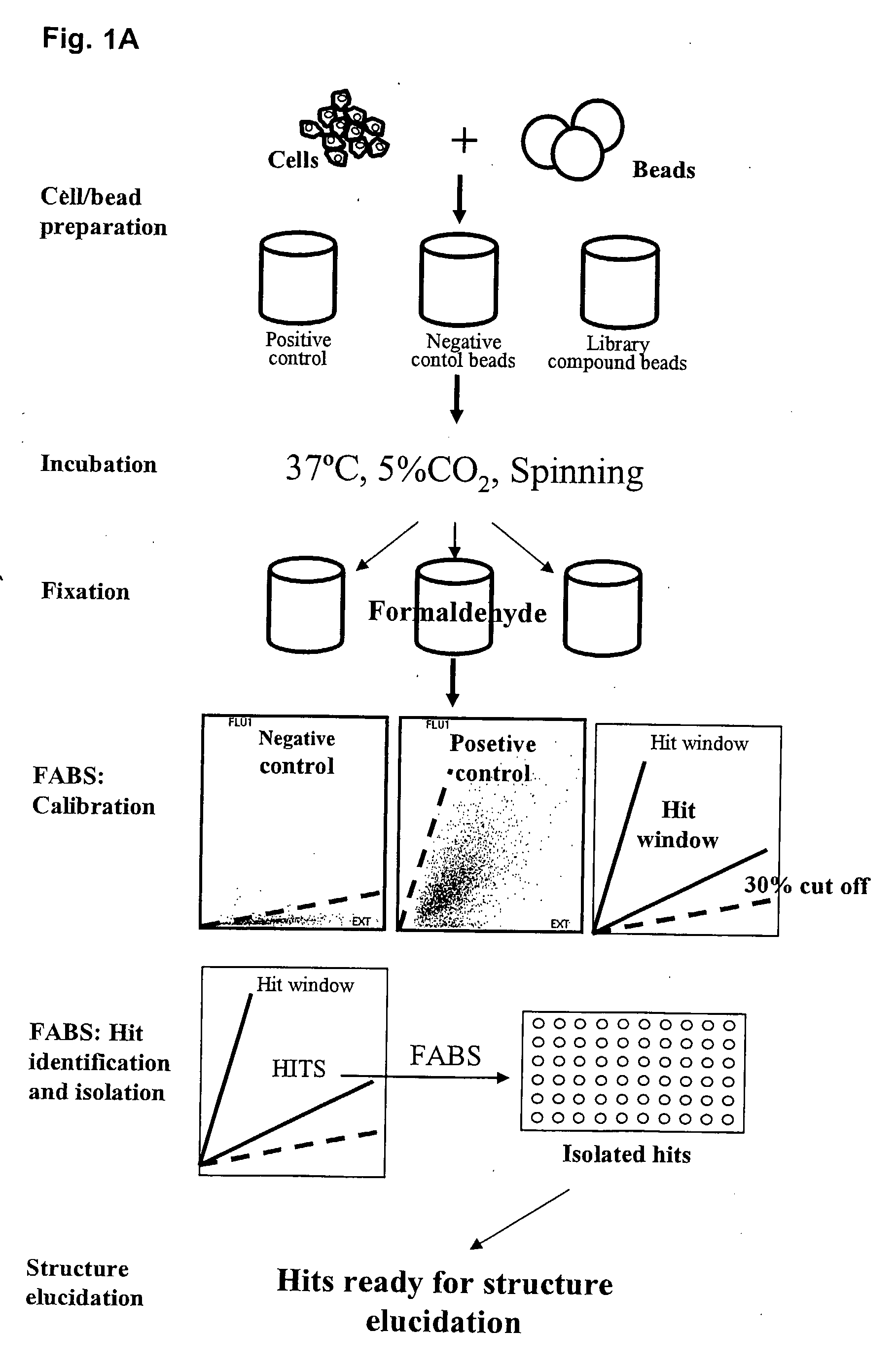
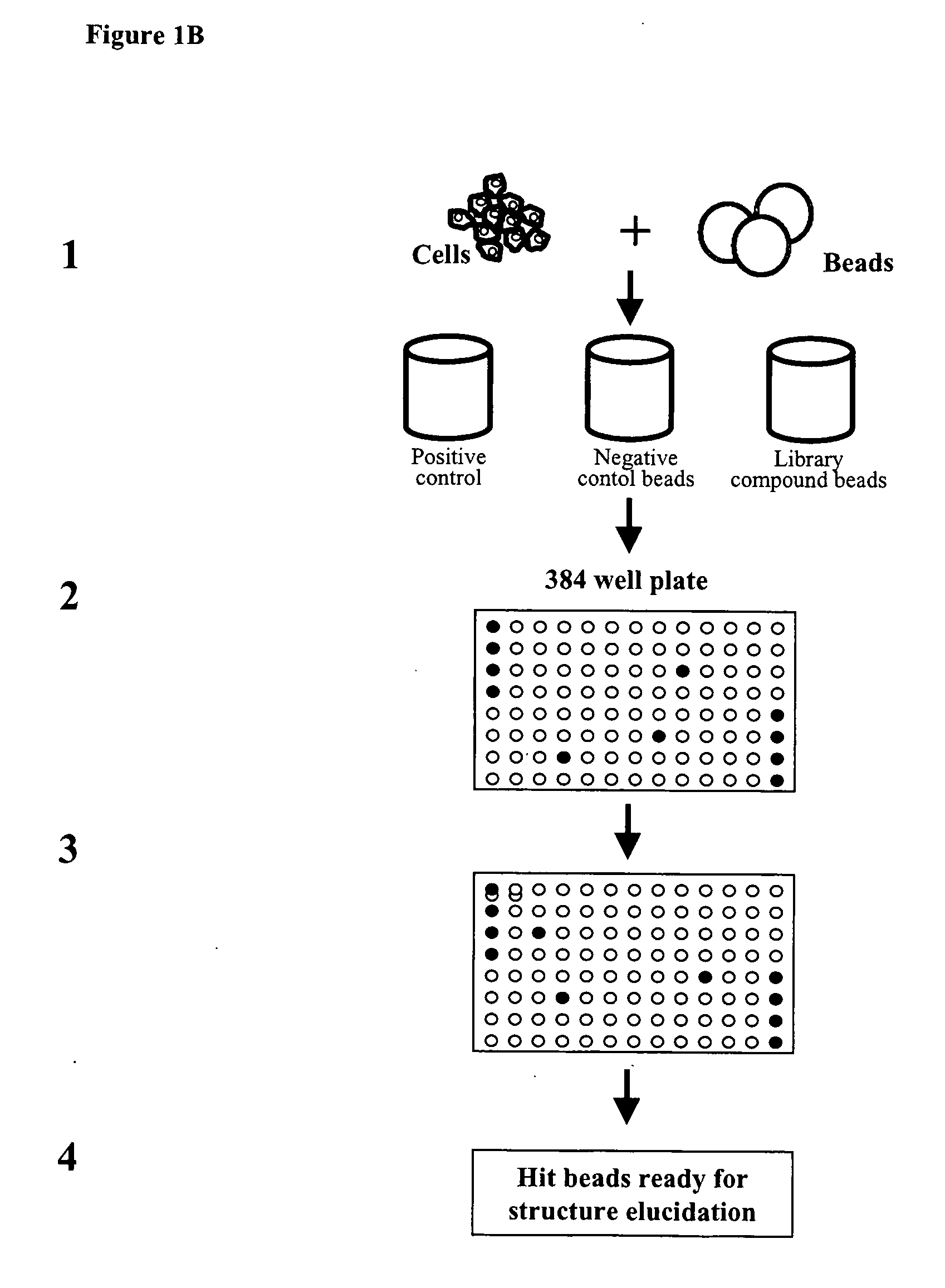
[0323]Approx. 100 adhesion peptide library beads were mixed with 1×10E6 cells (BHK, CHO, U2OS, Hek) in each well of a Falcon 12 well plate using 2 ml growth medium. The adhesion peptide library was prepared according to the general methods for solid phase peptide synthesis outlined above. The library consisted of heptamers of D-amino acids. The peptide library beads were PEGA beads each coupled to a potential adhesion peptide. The cells and beads were mixed gently every 15 min for 2 hrs. Supernatant with nonattached cells were removed and new growth medium added. Cells / beads were incubating for another 16 hrs. (37° C., 5% CO2). Cell adhesive beads were identified using a microscope with 10× objective and positive beads were transferred to a filter paper (to suck off medium). Peptides were identified by amino acid sequencing. Examples of useful peptides are given in table 2.
example 2b
Identification of an Adhesion Peptide with Low Absorption of Fluorescent Components from Growth Medium and High Adhesion Properties
[0324]An adhesion D-amino peptide library was synthesized (500.000 members) as described above in Example 2a and screened for low fluorescence / high adherence properties. This was done in 4 steps:
1) Selection of low fluorescent beads by Fluorescence Activated Bead Sorting (FABS).
[0325]The 500.000 member adhesion peptide library was FABSorted and 150.000 low fluorescent beads were isolated.
2) Selection of beads with good cell adhesion properties.
[0326]The 150.000 low fluorescent beads were incubated with GFP expressing U2OS cells followed by FABS sorting for high fluorescence (high cell adhesion). 536 beads were isolated.
3) Identification and isolation of beads with high Hek293 cell adherence properties.
[0327]The 536 beads were cleared for U2OS cells and incubated with GFP expressing Hek293 cells. 47 beads with high cell adhesion properties were isolated u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com