Methods to identify inhibitors of the unfolded protein response
a technology of unfolded protein and inhibitors, applied in the field of methods to identify inhibitors of unfolded protein response, can solve the problems of cell death, increased expression, and inability to account for the full repertoire of changes of hif activation alone, and achieve the effect of inhibiting tumor growth in vivo
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
Involvement of XBP-1 in Hypoxia and Tumor Growth
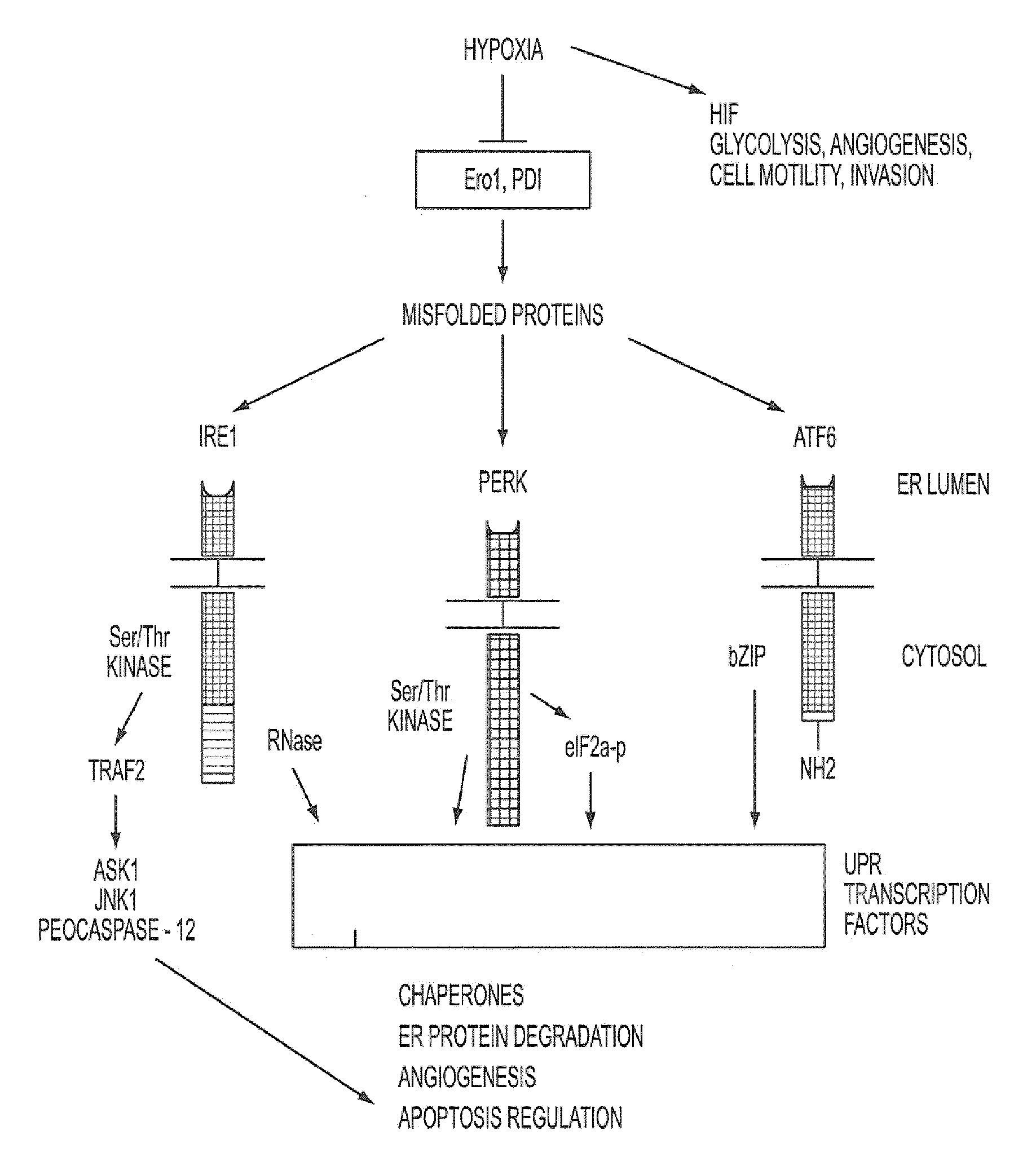
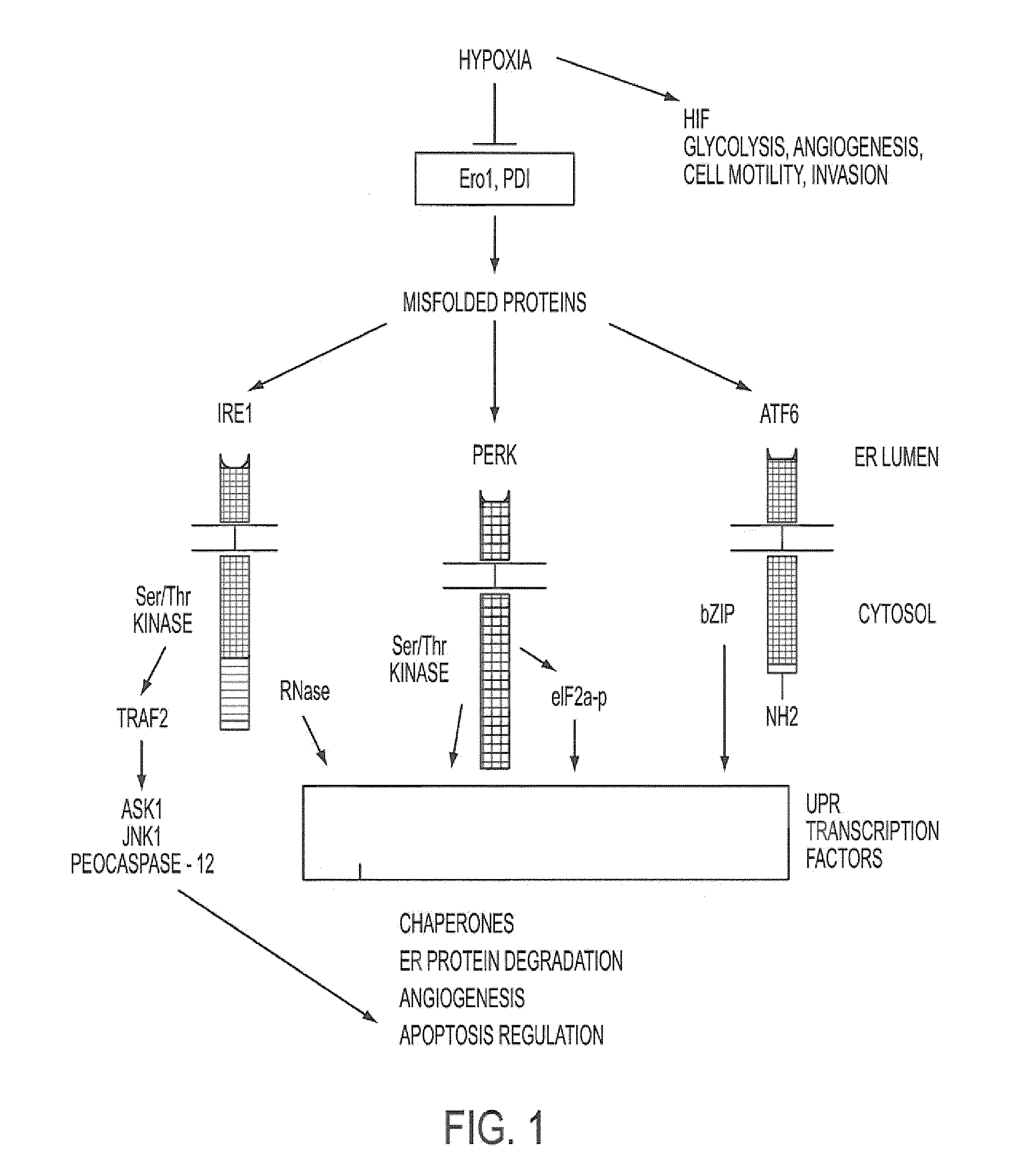
[0107]We have demonstrated that UPR related genes represent a major class of genes that are transcriptionally induced under hypoxia, that XBP-1 is activated during hypoxia in a HIF-1 independent manner, and that cell survival and apoptosis under hypoxia was mediated by XBP-1 (Romero L., et al. Cancer Res. 64:5943-5947, 2004). We have demonstrated that XBP-1 is essential for tumor growth. We implanted spontaneously transformed XBP-1 wild-type and knockout mouse embryonic fibroblasts (MEFs) as tumor xenografts into SCID mice and found that XBP-1 knockout MEFs were completely unable to grow as tumors. Furthermore, tumor growth was dependent upon the spliced form of XBP-1. We transfected spliced XBP-1 (XBP1s) into XBP-1 knockout MEFs and were able to restore the growth rate of these tumors back to that of the wild-type cells. We also transfected a mutant form of unspliced XBP-1 (XBP1u) in which the splice site was deleted. Transfection of ...
example 2
Identification of Inhibitors of XBP-1 Splicing
[0112]A high throughput screen for small molecule inhibitors of IRE1 activity was developed as detailed below. The sequence for XBP-1 is described in, for example, Liou, H-C. et al. Science 247:1581-1584, 1990; and Yoshimura, T. et al. EMBO J. 9:2537-2542, 1990. The amino acid sequence for unspliced XBP-1 protein is provided in SEQ ID NO: 1, with corresponding cDNA sequence being provided in SEQ ID NO: 3. The amino acid sequence for the spliced form is provided in SEQ ID NO: 2.
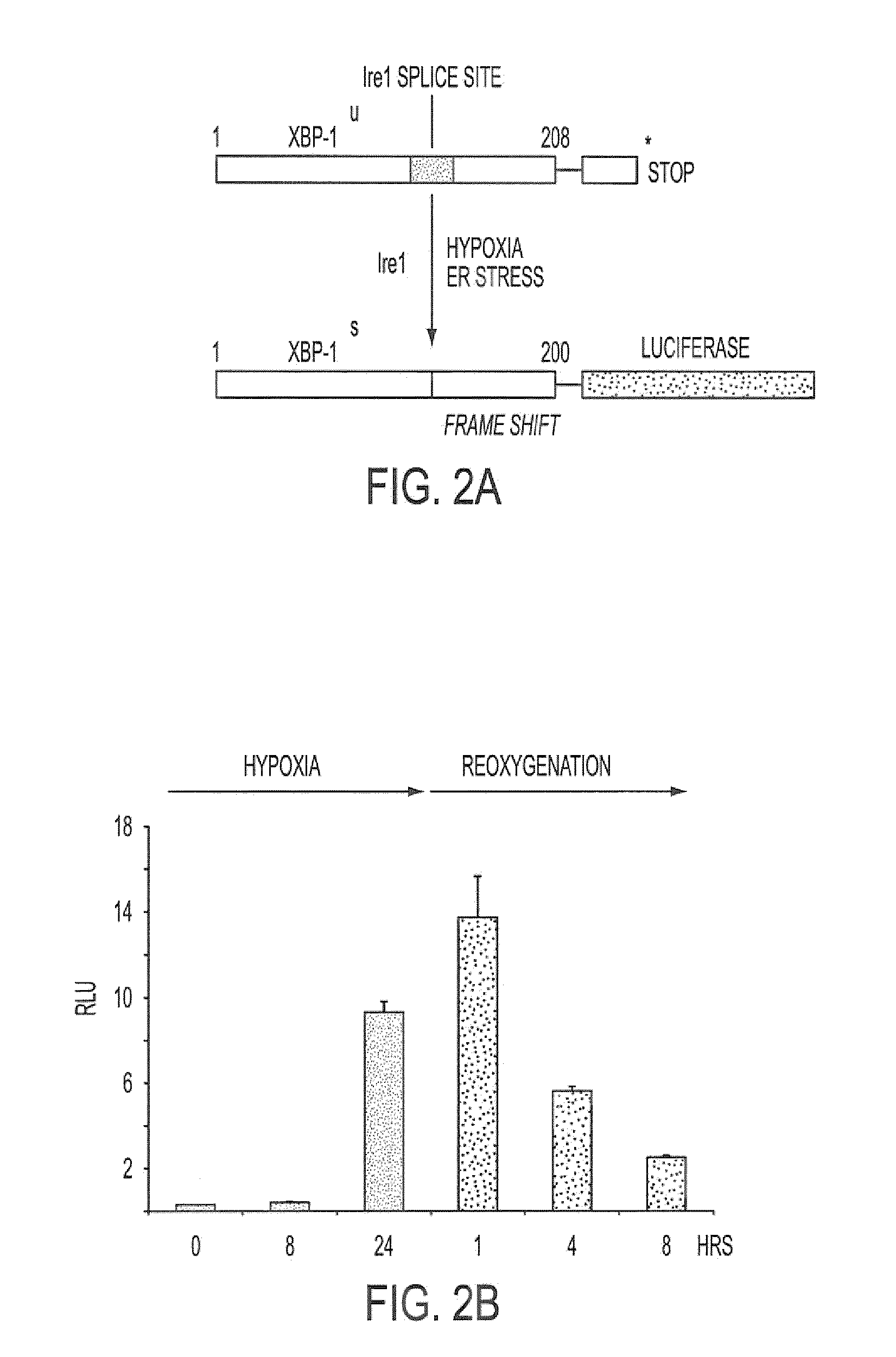
[0113]As shown in FIG. 2A, we developed a reporter construct in which luciferase was fused downstream and in frame with the unspliced form of XBP-1, containing the IRE-1 splice site. In the unspliced form, no luciferase is translated because of an endogenous stop codon. However, during hypoxia and ER stress, a 26 nt sequence is spliced out by IRE1 resulting in a frame-shift and read-through of the stop codon (Iwawaki et al., Nat. Med. 10:98-102, 2004). This results...
example 3
Inhibition of XBP-1 Splicing in Tumors by Inhibitors of IRE1 Activity
[0125]Several nude mice were implanted with HT1080 cells stably expressing a XBP-1s-luciferase construct and XBP-1 activation was examined using bioluminescence imaging. Imaging was performed using the In Vivo Imaging System (IVIS, Xenogen Corporation, Alameda, Calif.) in the Stanford Center for Innovation in In Vivo Imaging (SCI3). This device consists of a cooled CCD camera mounted on a light-tight specimen chamber. In these experiments, two different potential irestatins (3281 & 5500) were injected IP into nude mice implanted with HT1080 stably expressing XBP1s-luciferase (described in FIG. 2A). We estimated that injecting mice at a concentration of 50 mg / kg (no apparent toxicity) was within a 10-fold range of the in vitro drug concentrations used (assuming uniform distribution and ignoring excretion / metabolism) for the above described cell culture assays.
[0126]As shown in FIGS. 9A-D, XBP-1 splicing activity was...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
| total volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com