Spinosyn-producing polyketide synthases
a polyketide synthase and spinosyn technology, applied in the field of spinosyn-producing polyketide synthases, can solve the problem that not all manipulations of pks genes produce the targeted new analogues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of pCJR81
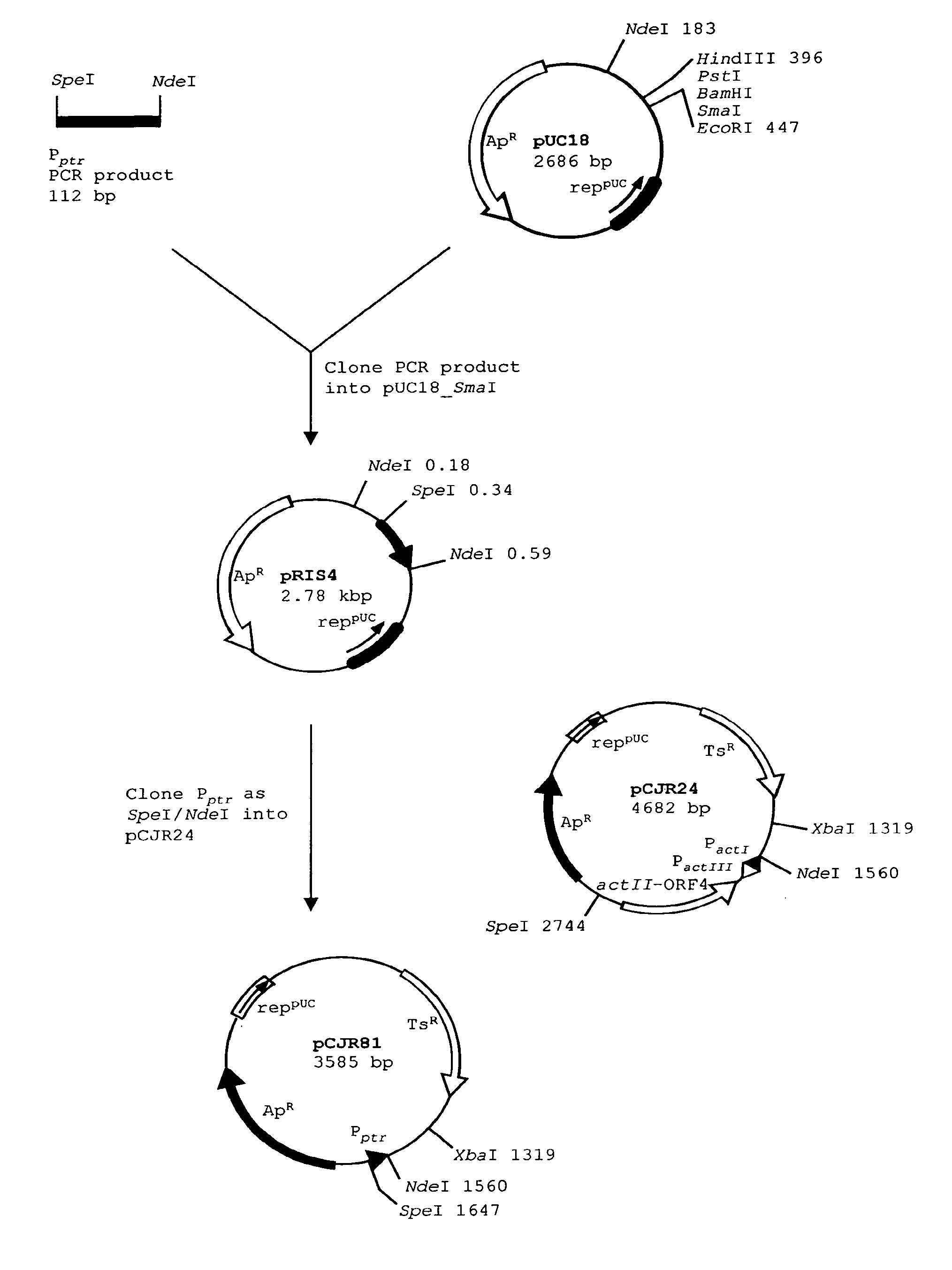
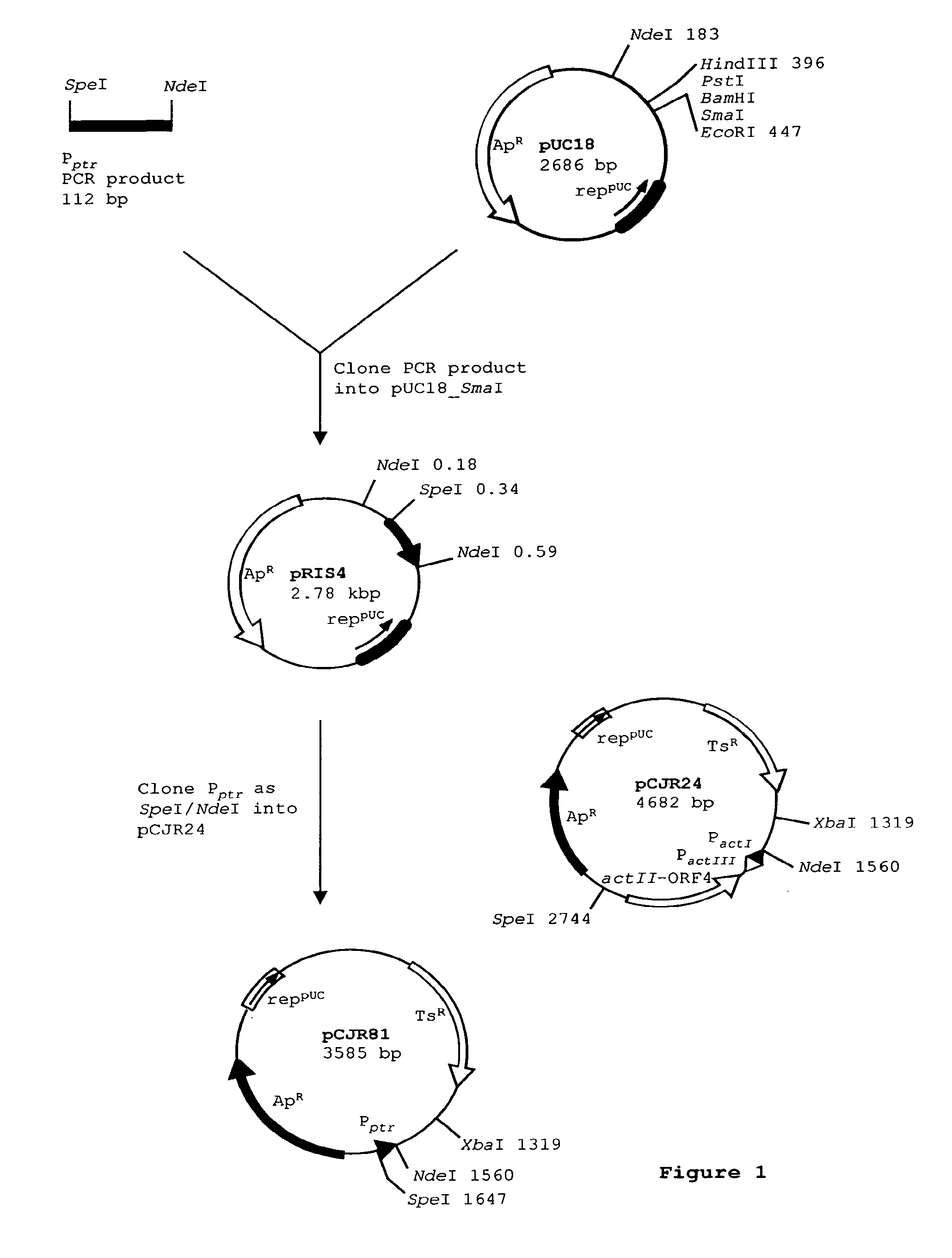
[0108]See FIG. 1. Plasmid pCJR81 is a vector for expression of polyketide genes under the promoter for resistance to pristinamycin. It was constructed as follows:
[0109]Two overlapping oligos were designed to perform a PCR reaction in which they act both as primers and template. They were designed to introduce an NdeI restriction site incorporating the ATG start codon, such that genes can be cloned with optimal spacing from the ribosome binding site. A SpeI restriction site was incorporated to facilitate further cloning. The oligos are PRIS1 (SEQ ID NO:1) and PRIS2(SEQ ID NO:2).
[0110]Amplification to obtain the promoter fragment was performed with Pwo thermostable DNA polymerase using the manufacturer's conditions. The 112 bp fragment was phosphorylated with T4 polynucleotide kinase, and cloned into commercially-available pUC18 digested with SmaI and dephosphorylated. Plasmids containing inserts were sequenced. One plasmid containing the correct sequence was des...
example 2
Construction of Plasmids for Expression from S. spinosa
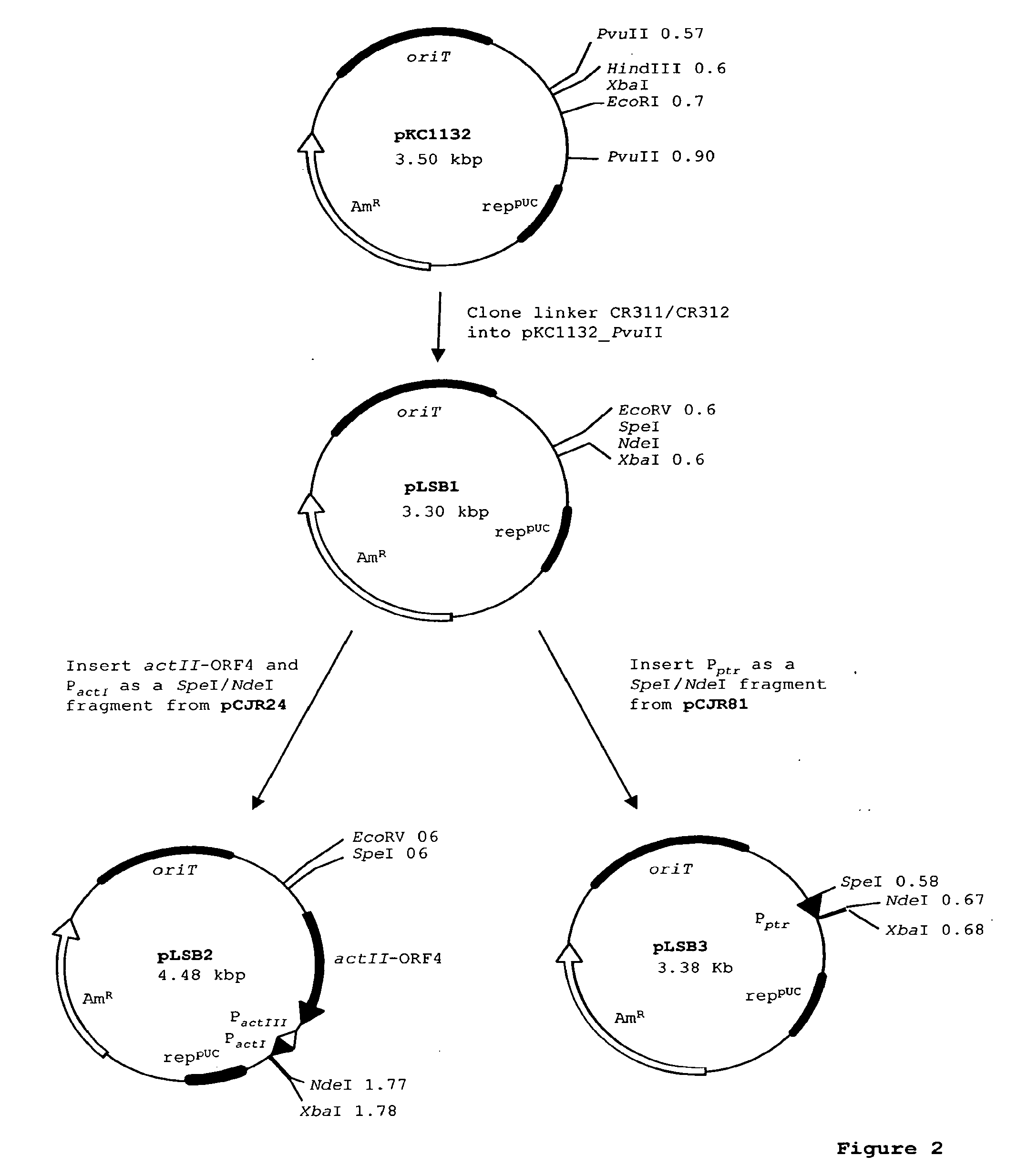
[0112]See FIG. 2. Plasmids pLSB2 and pLSB3 were constructed for expression of polyketide genes or accessory genes in S. spinosa. Plasmid pLSB2 contains the actI romoter (PactI) and its cognate activator, actII-ORF4. Plasmid pLSB3 contains the promoter for resistance to pristinamycin (Pptr). These plasmids were constructed as follows:
[0113]Plasmid pKC1132 (Bierman et al. 1992) contains an origin of transfer (oriT), and an apramycin resistance marker for selection in both E. coli and actinomycetes. It can therefore be used for DNA manipulations in E. coli, and permit the final plasmids to be introduced into S. spinosa by conjugation. The polylinker of pKC1132 was replaced by a linker of two oligos CR311 (SEQ ID NO:4) and CR312 (SEQ ID NO:5) in order to provide appropriate EcoRV / SpeI / NdeI / XbaI restriction sites.
[0114]Plasmid pKC1132 was digested with PvuII and the ends dephosphorylated with shrimp alkaline phosphatase. The oligos ...
example 3
Construction of a Vector to Incorporate the Loading Module of the Erythromycin Polyketide Synthase into the Spinosyn Polyketide Synthase
[0115]See FIG. 3. The vector to incorporate the loading module of the erythromycin polyketide synthase into the spinosyn polyketide synthase contains the erythromycin loading module (AT0ACP0), followed by a region of the first module of the spinosyn PKS to provide homology for integration. The vector is designated pLSB62 and was constructed as follows.
[0116]The erythromycin loading module was amplified by PCR using pCJR26 (Rowe et al. 1998) as the template, and oligos SP28 (SEQ ID NO:6) and SP29 (SEQ ID NO:7). SP28 incorporates an NdeI site at the start codon of the ery sequence, and SP29 incorporates an NheI site at the beginning of the KS1 domain.
[0117]The PCR fragment was phosphorylated, gel-purified and cloned into pUC18 which had been previously digested with SmaI and dephosphorylated. Clones were screened for the presence of inserts and sequen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com