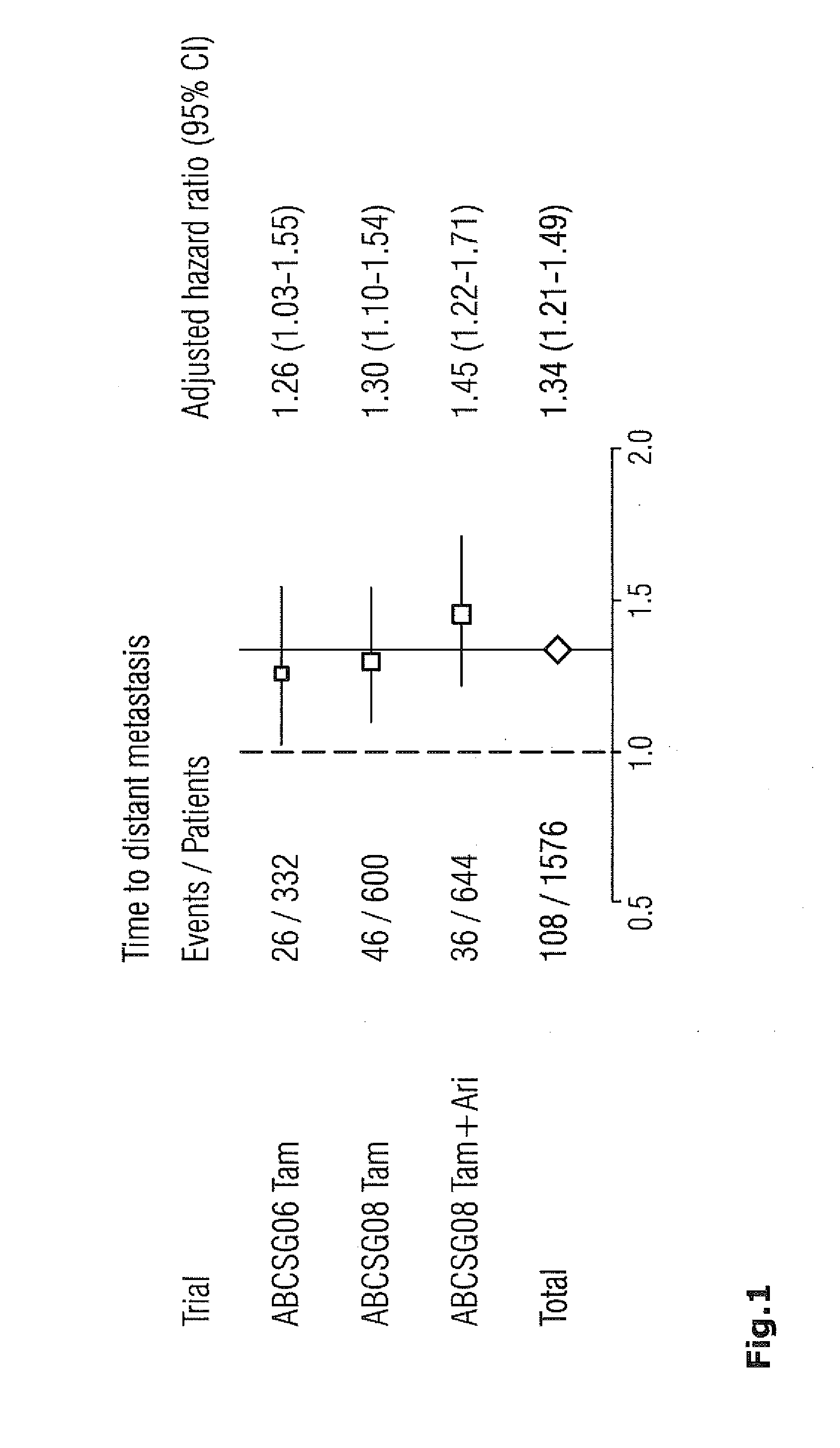
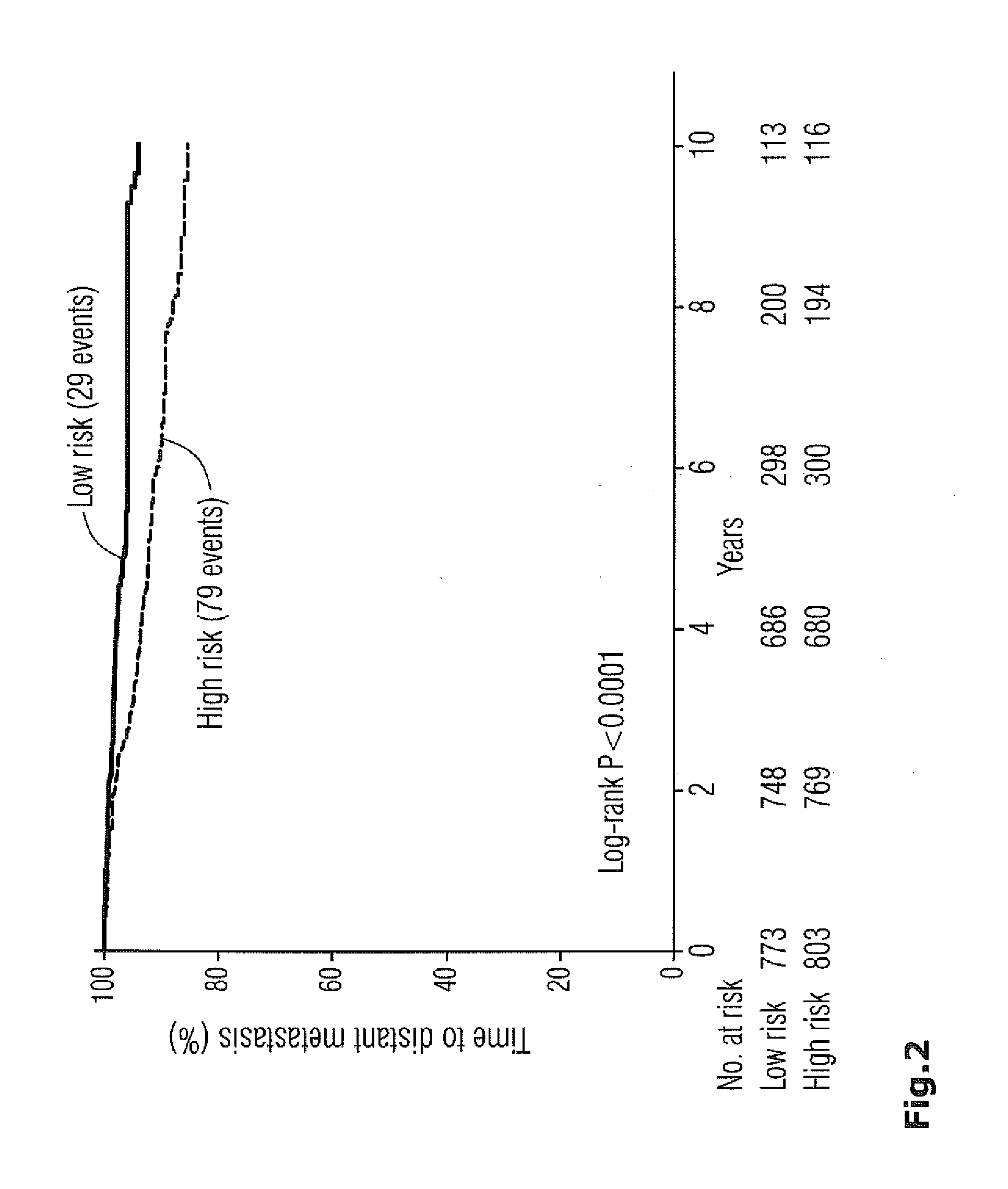
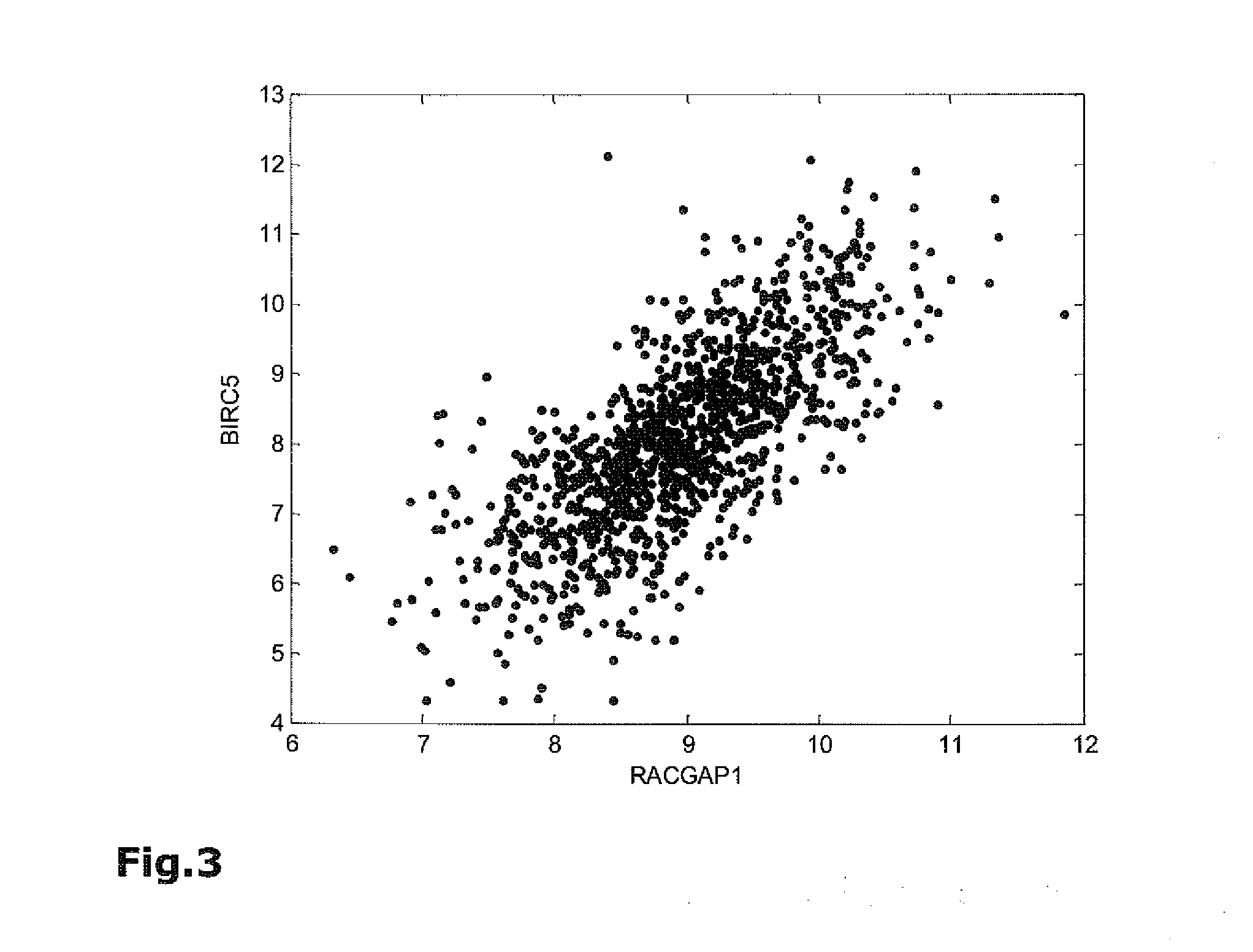
Method for breast cancer recurrence prediction under endocrine treatment
a breast cancer and endocrine treatment technology, applied in the field of breast cancer recurrence prediction under endocrine treatment, can solve the problems of high risk score, severe impairing patient quality of life, and inability to show significant benefit of adding cytotoxic agents alone against tamoxifen plus chemotherapy,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example algorithm t5
[0105]Algorithm T5 is a committee of four members where each member is a linear combination of two genes. The mathematical formulas for T5 are shown below; the notation is the same as for T1. T5 can be calculated from gene expression data only.
riskMember1=0.434039[0.301…0.567]*(0.939*BIRC5-3.831)-0.491845[-0.714…-0.270]*(0.707*RBBP8-0.934)riskMember2=0.488785[0.302…0.675]*(0.794*UBE2C-1.416)-0.374702[-0.570…-0.179]*(0.814*IL6ST-5.034)riskMember3=-0.39169[-0.541…-0.242]*(0.674*AZGP1-0.777)+0.44229[0.256…0.628]*(0.891*DHCR7-4.378)riskMember4=-0.377752[-0.543…-0.212]* (0.485*MGP+4.330)-0.177669[-0.267…-0.088]*(0.826*STC2-3.630)risk=riskMember1+riskMember2+riskMember3+riskMember4
[0106]Coefficients on the left of each line were calculated as COX proportional hazards regression coefficients, the numbers in squared brackets denote 95% confidence bounds for these coefficients. In other words, instead of multiplying the term (0.939*BIRC5−3.831) with 0.434039, it may be multiplied with any co...
example algorithm t1
[0113]Algorithm T1 is a committee of three members where each member is a linear combination of up to four variables. In general variables may be gene expressions or clinical variables. In T1 the only non-gene variable is the nodal status coded 0, if patient is lymph-node negative and 1, if patient is lymph-node-positive. The mathematical formulas for T1 are shown below.
riskMember1=+0.193935[0.108…0.280]*(0.792*PVALB-2.189)-0.240252[-0.400…-0.080]*(0.859*CDH1-2.900)-0.270069[-0.385…-0.155]*(0.821*STC2-3.529)+1.2053[0.534…1.877]*nodalStatusriskMember2=-0.25051[-0.437…-0.064]*(0.558*CXCL12+0.324)-0.421992[-0.687…-0.157]*(0.715*RBBP8-1.063)+0.148497[0.029…0.268]*(1.823*NMU-12.563)+0.293563[0.108…0.479]*(0.989*BIRC5-4.536)riskMember3=+0.308391[0.074…0.543]*(0.812*AURKA-2.656)-0.225358[-0.395…-0.055]*(0.637*PTGER3+0.492)-0.116312[-0.202…-0.031]*(0.724*PIP+0.985)risk=+riskMember1+riskMember2+riskMember3
[0114]Coefficients on the left of each line were calculated as COX proportional hazards...
example algorithm t4
[0115]Algorithm T4 is a linear combination of motifs. The top 10 genes of several analyses of Affymetrix datasets and PCR data were clustered to motifs. Genes not belonging to a cluster were used as single gene-motifs. COX proportional hazards regression coefficients were found in a multivariate analysis.
[0116]In general motifs may be single gene expressions or mean gene expressions of correlated genes. The mathematical formulas for T4 are shown below.
prolif=((0.84 [0.697 . . . 0.977]*RACGAP1−2.174)+(0.85 [0.713 . . . 0.988]*DHCR7−3.808)+(0.94 [0.786 . . . 1.089]*BIRC5−3.734)) / 3
motiv2=((0.83 [0.693 . . . 0.96]*IL6ST−5.295)+(1.11 [0.930 . . . 1.288]*ABAT−7.019)+(0.84 [0.701 . . . 0.972]*STC2−3.857)) / 3
ptger3=(PTGER3*0.57 [0.475 . . . 0.659]+1.436)
cxcl12=(CXCL12*0.53 [0.446 . . . 0.618]−0.847)
pvalb=(PVALB*0.67 [0.558 . . . 0.774]−0.466)
[0117]Factors and offsets for each gene denote a platform transfer from PCR to Affymetrix: The variables RACGAP1, DHCR7, . . . denote PCR-based expressi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| size | aaaaa | aaaaa |
| weight | aaaaa | aaaaa |
| imaging methods | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com