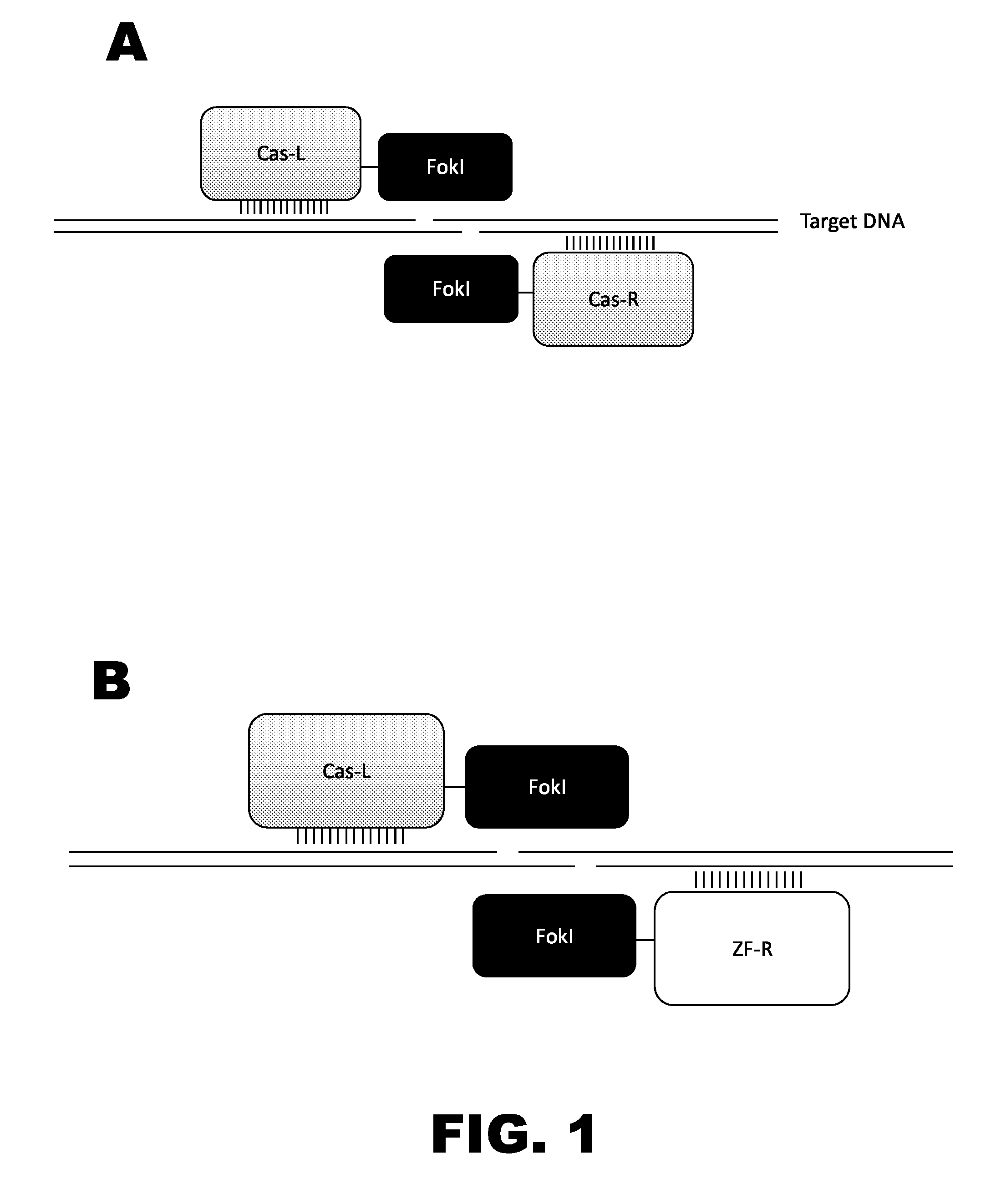
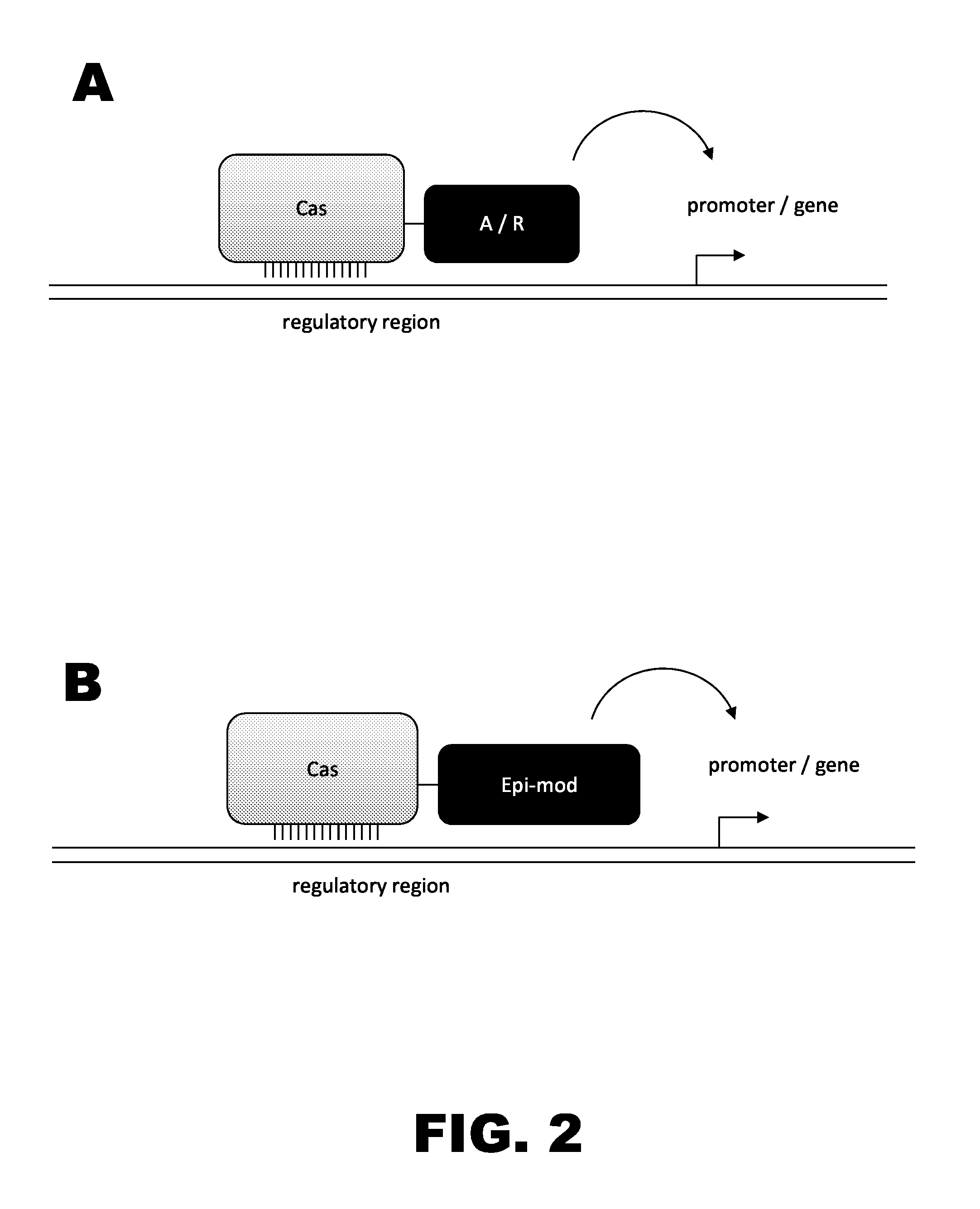
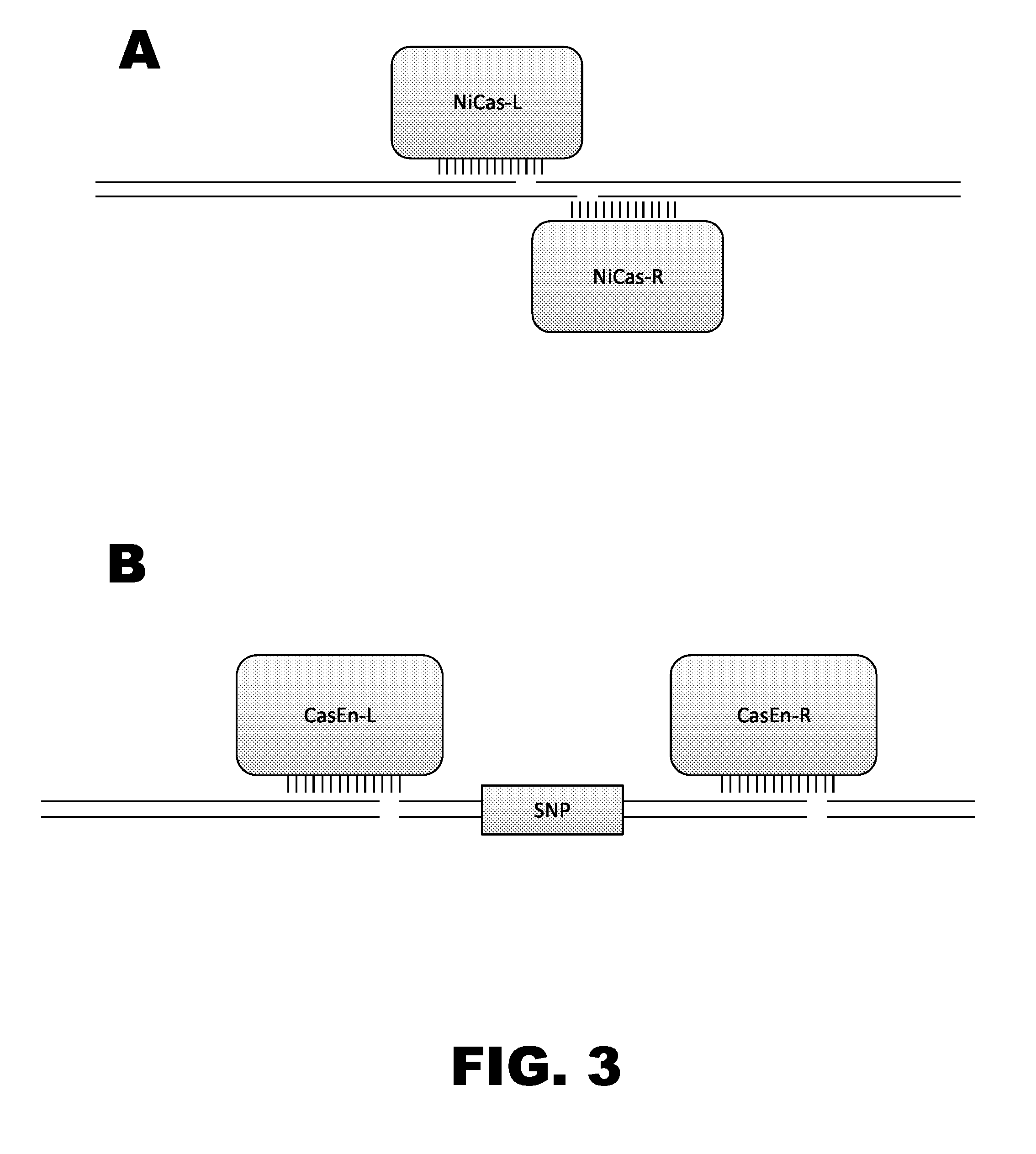
Crispr-based genome modification and regulation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Modification of Cas9 Gene for Mammalian Expression
[0160]A Cas9 gene from Streptococcus pyogenes strain MGAS15252 (Accession number YP—005388840.1) was optimized with Homo sapiens codon preference to enhance its translation in mammalian cells. The Cas9 gene also was modified by adding a nuclear localization signal PKKKRKV (SEQ ID NO:1) at the C terminus for targeting the protein into the nuclei of mammalian cells. Table 1 presents the modified Cas9 amino acid sequence, with the nuclear localization sequence underlined. Table 2 presents the codon optimized, modified Cas9 DNA sequence.
TABLE 1Modified Cas9 Amino Acid SequenceMDKKYSIGLDIGTNSVGWAVITDDYKVPSKKFKVLGNTDRHSIKKNLIGALLFGSGETAEATRLKRTARRRYTRRKNRICYLQEIFSNEMAKVDDSFFHRLEESFLVEEDKKHERHPIFGNIVDEVAYHEKYPTIYHLRKKLADSTDKADLRLIYLALAHMIKFRGHFLIEGDLNPDNSDVDKLFIQLVQIYNQLFEENPINASRVDAKAILSARLSKSRRLENLIAQLPGEKRNGLFGNLIALSLGLTPNFKSNFDLAEDAKLQLSKDTYDDDLDNLLAQIGDQYADLFLAAKNLSDAILLSDILRVNSEITKAPLSASMIKRYDEHHQDLTLLKALVRQQLPEKYKEIFFDQSKNGYAGYIDGGAS...
example 2
Targeting Cas9
[0162]The adeno-associated virus integration site 1 (AAVS1) locus was used as a target for Cas9-mediated human genome modification. The human AAVS1 locus is located in intron 1 (4427 bp) of protein phosphatase 1, regulatory subunit 12C (PPP1R12C). Table 3 presents the first exon (shaded gray) and the first intron of PPP1R12C. The underlined sequence within the intron is the targeted modification site (i.e., AAVS1 locus).
TABLE 3First Exon and Intron of PPP1R12C (5'-3')GCCCGGCGTCTCCCGGGGCCAGGTCCACCCTCTGCTGCGCCACCTGGGGCATCCTCCTTCCCCGTTGCCAGTCTCGATCCGCCCCGTCGTTCCTGGCCCTGGGCTTTGCCACCCTATGCTGACACCCCGTCCCAGTCCCCCTTACCATTCCCCTTCGACCACCCCACTTCCGAATTGGAGCCGCTTCAACTGGCCCTGGGCTTAGCCACTCTGTGCTGACCACTCTGCCCCAGGCCTCCTTACCATTCCCCTTCGACCTACTCTCTTCCGCATTGGAGTCGCTTTAACTGGCCCTGGCTTTGGCAGCCTGTGCTGACCCATGCAGTCCTCCTTACCATCCCTCCCTCGACTTCCCCTCTTCCGATGTTGAGCCCCTCCAGCCGGTCCTGGACTTTGTCTCCTTCCCTGCCCTGCCCTCTCCTGAACCTGAGCCAGCTCCCATAGCTCAGTCTGGTCTATCTGCCTGGCCCTGGCCATTGTCACTTTGCGCTGCCCTCCTCTCGCCCCCGAG...
example 3
Preparation of Donor Polynucleotide to Monitor Genome Modification
[0164]Targeted integration of a GFP protein into the N terminus of PPP1R12C was used to monitor Cas9-mediated genome modification. To mediate integration by homologous recombination a donor polynucleotide was prepared. The AAVS1-GFP DNA donor contained a 5′ (1185 bp) AAVS1 locus homologous arm, an RNA splicing receptor, a turbo GFP coding sequence, a 3′ transcription terminator, and a 3′ (1217 bp) AAVS1 locus homologous arm. Table 5 presents the sequences of the RNA splicing receptor and the GFP coding sequence followed by the 3′ transcription terminator. Plasmid DNA was prepared by using GenElute Endotoxin-Free Plasmid Maxiprep Kit (Sigma).
TABLE 5Sequences in the AAVS1-GFP DNA donor sequenceSEQID5′-3′ SequenceNO:RNA splicingCTGACCTCTTCTCTTCCTCCCACAG15receptorGFP codingGCCACCATGGACTACAAAGACGATGACGAC16sequence andAAGGTCGACTCTAGAGCTGCAGAGAGCGACtranscription GAGAGCGGCCTGCCCGCCATGGAGATCGAGterminatorTGCCGCATCACCGGCACCCTGAA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com