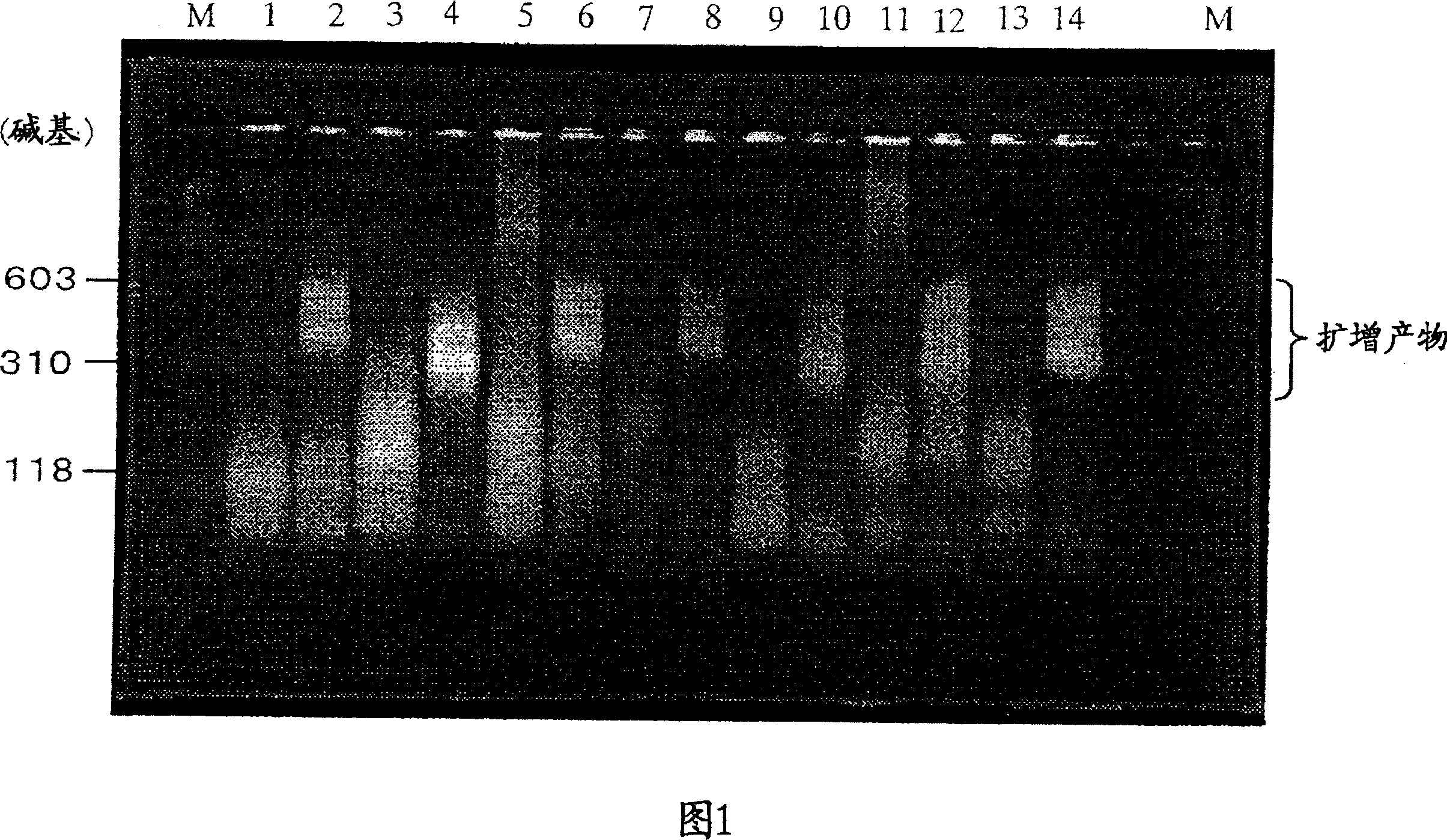
Method of detecting micrometastasis
A technology for micro-transfer and production testing, applied in biochemical equipment and methods, microbial determination/inspection, organic chemistry, etc., can solve the problems of delayed test results and difficult mRNA processing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Carcinoembryonic antigen (CEA) RNA is specifically amplified by using the combination of oligonucleotide primers of the present invention. CEA RNA was prepared and isolated by in vitro transcription of double-stranded DNA containing the nucleotide sequence of the CEA gene (National Center for Biotechnology Information accession number: M29540). In this embodiment, specific sequences refer to the transcripts listed in Table 1 below.
[0042] (1) CEA RNA (2109-mer) as a sample was quantified by measuring the UV absorbance at 260nm, and the RNA diluent (10mM Tris-HCl (pH 8.0), 0.1mM EDTA, 0.5U / μl RNase inhibitor, 5.0mM DTT) diluted to 1.0×10 4 copies / 5 μl. Dilutions were used as controls (negative).
[0043] (2) Add 20.8 μl of reaction solution composed of the following components into a 0.5 ml PCR tube (GeneAmp thin-walled reaction tube, Perkin Elmer), and add 5.0 μl of the above-mentioned CEA RNA sample.
[0044] Composition of reaction solution (final concentration ...
Embodiment 2
[0071] Preparation of fluorescent intercalating dye-labeled oligonucleotide probes
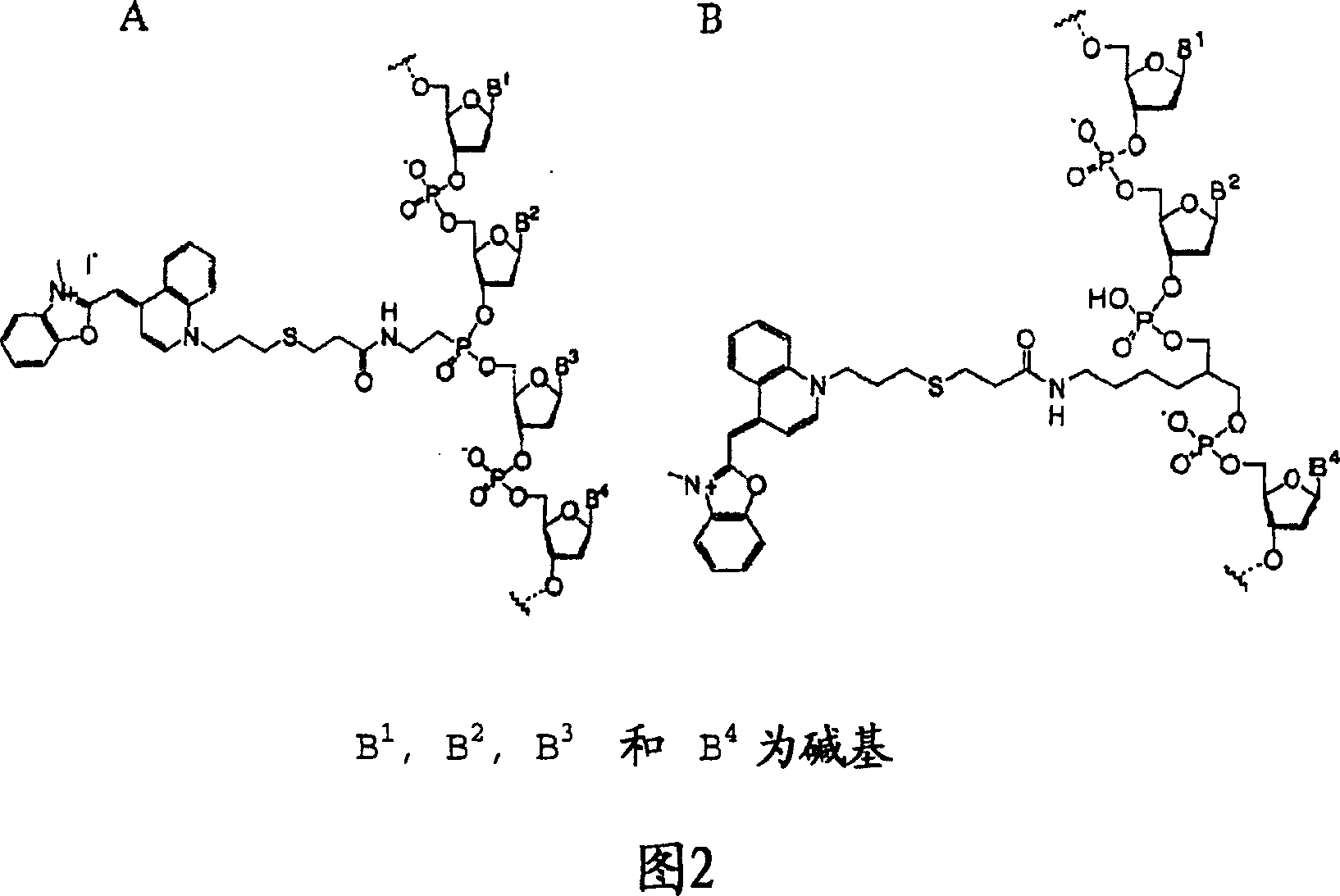
[0072] The 20-base oligonucleotide having the sequence shown in SEQ ID NO: 19 is modified by C 15 The linker uses Label-ON reagent (Clontech) to connect the amino group between the 5th nucleotide (G) and the 6th nucleotide (A) at its 5' end, and its 3' end is further modified with biotin . Oxazole yellow (oxazole yellow) is a commonly used fluorescent intercalating dye, which can be used as a marker to prepare oxazole yellow-labeled oligonucleotide probes (SEQ ID NO: 19) (Fig. 2B, Ishiguro T (1996) Nucleic Acids Res. 24(24) 4992-4997).
Embodiment 3
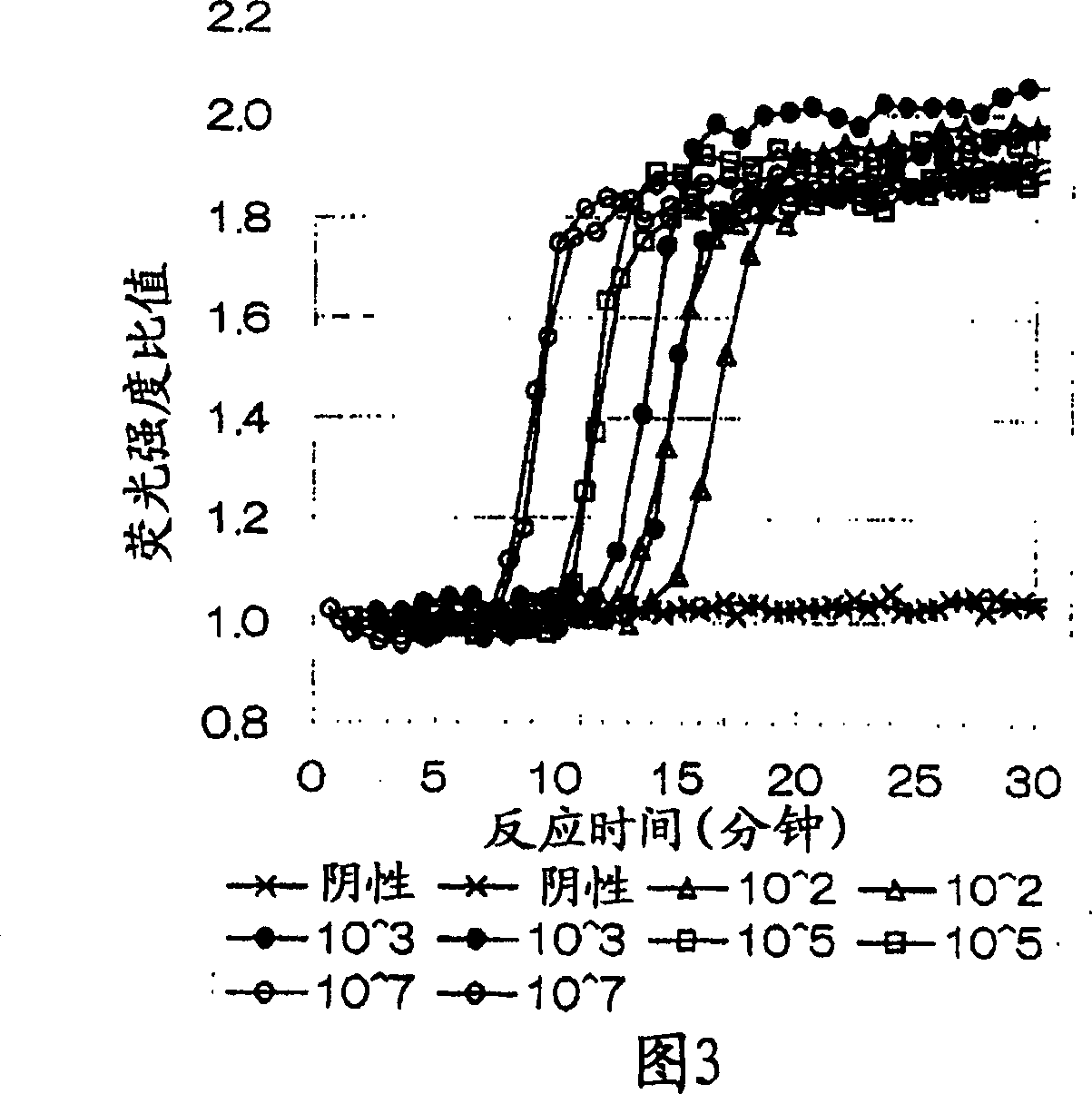
[0074] The combination of oligonucleotide primers in the present invention is used to detect CEA RNA with different initial copy numbers.
[0075] Sample CEA RNA (2109-mer) was quantified by measuring UV absorbance at 260nm, using RNA diluent (10mM Tris-HCl (pH 8.0), 0.1mM EDTA, 0.5U / μl RNase inhibitor, 5.0mM DTT) to Dilute it to 1.0 x 10 7 -10 2 Copies / 5 μl, the dilution was used as a control (negative).
[0076] 20.0 μl of the reaction solution with the following components was added to a 0.5 ml PCR tube (Gene Amp thin-walled reaction tube, Perkin Elmer), and 5.0 μl of the above-mentioned CEA RNA sample was added.
[0077] Composition of the reaction solution (final concentration in the reaction solution after adding the enzyme solution)
[0078] 60.0mM Tris-HCl buffer (pH 8.6)
[0079] 17.0mM magnesium chloride
[0080] 100.0mM potassium chloride
[0081] 1.0mM DTT
[0082] 0.25mM dATP, dCTP, dGTP, dTTP
[0083] 3.0mM ATP, CTP, UTP
[0084] 2.25mM GTP
[0085] 3.6...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com