Method of qualitative and quantitative analysis for algae
A technology for analyzing algae and probes, which is applied in the field of qualitative and quantitative analysis of algae, and can solve problems such as lack of microalgae, large fluctuations in test results, and difficult operations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
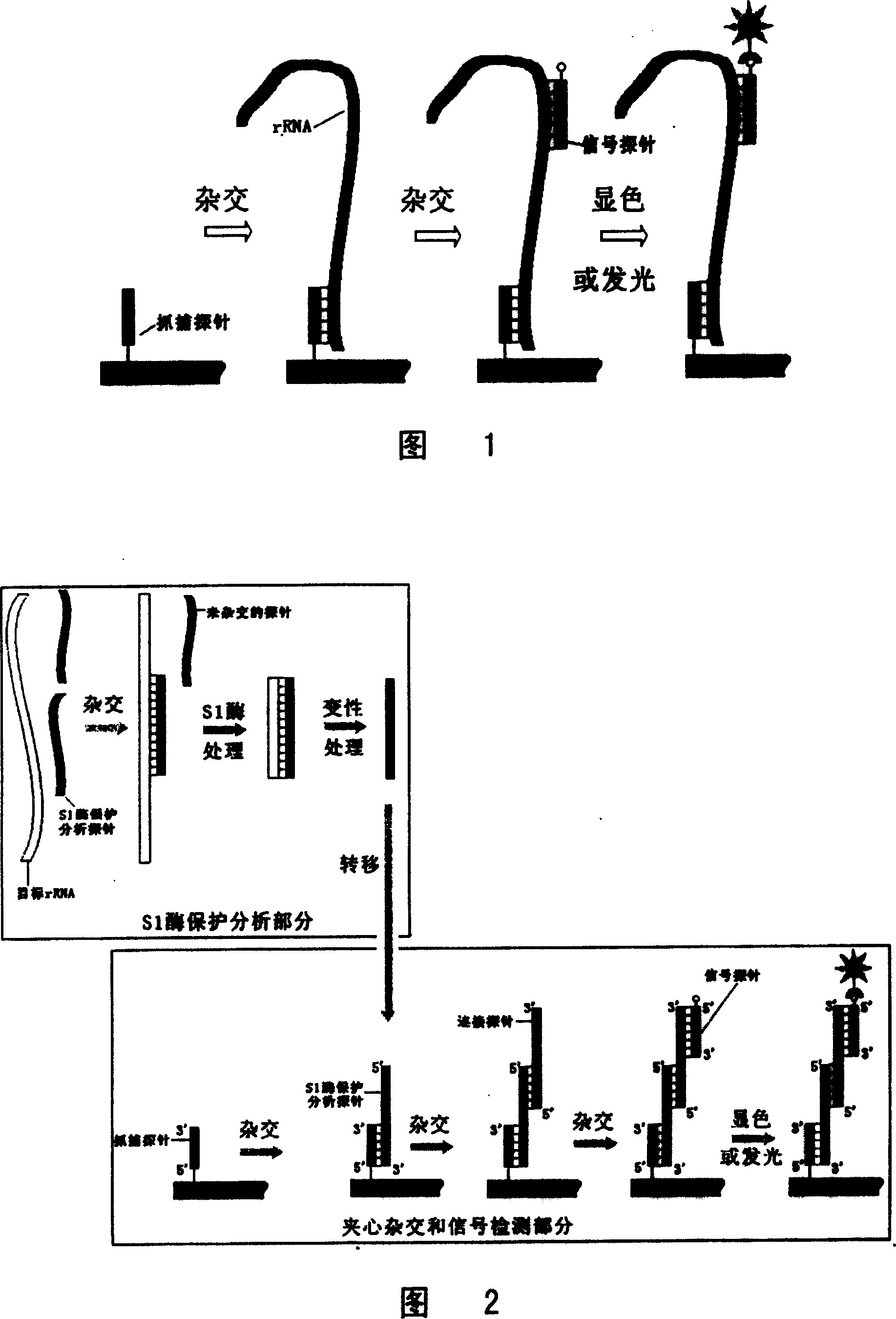
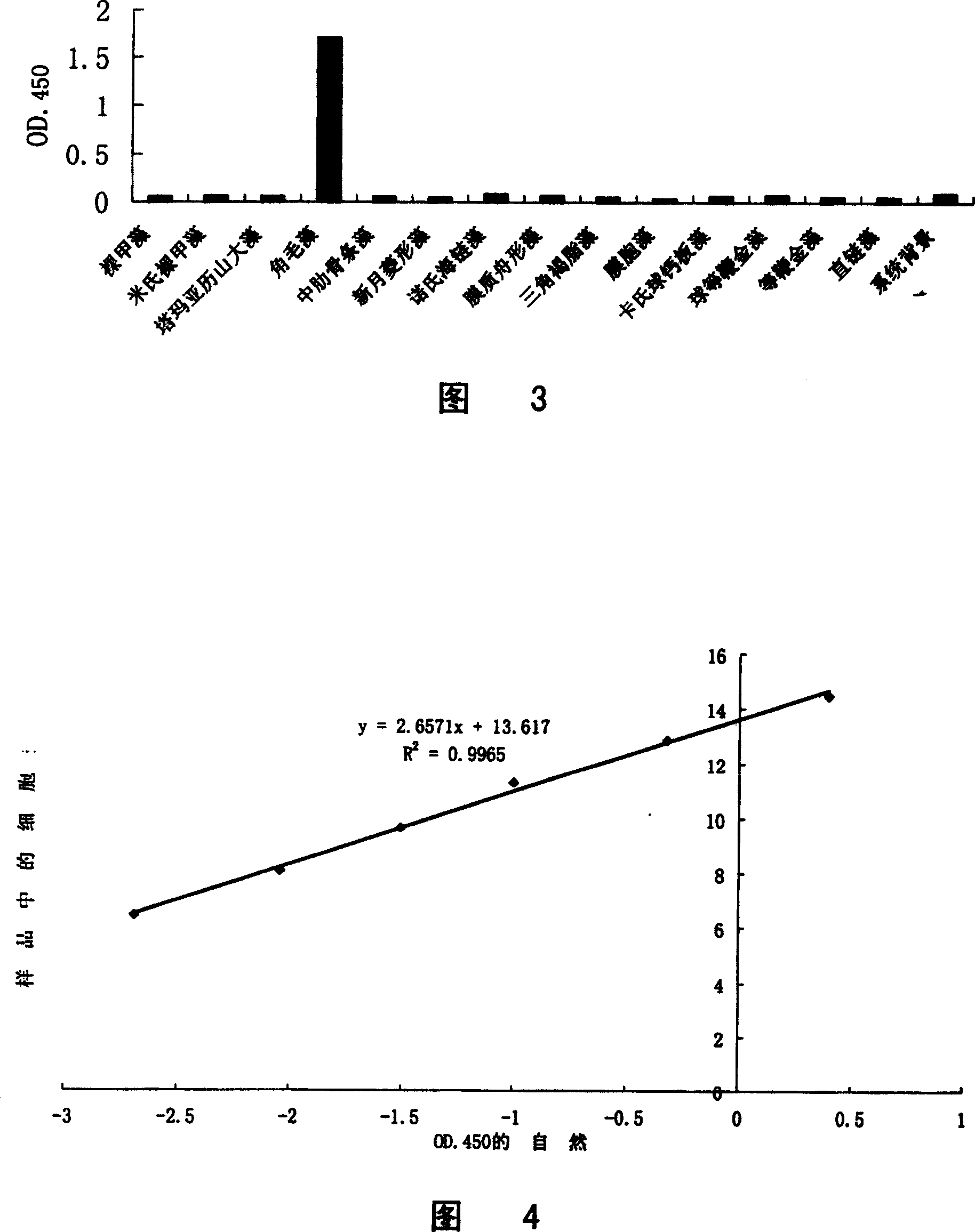
[0070] Example 1: Qualitative and quantitative analysis of Chaetoceros using S1 enzyme protection analysis for rRNA and sandwich hybridization technology
[0071] In this example, the S1 enzyme protection analysis technique and the sandwich hybridization technique are combined to realize the qualitative and quantitative analysis of the marine red tide organism Chaetoceros sp., and the steps are as follows:
[0072] 1.1 Search for the specific region of Chaetoceros rRNA and the design and synthesis of S1 enzyme protection analysis probe
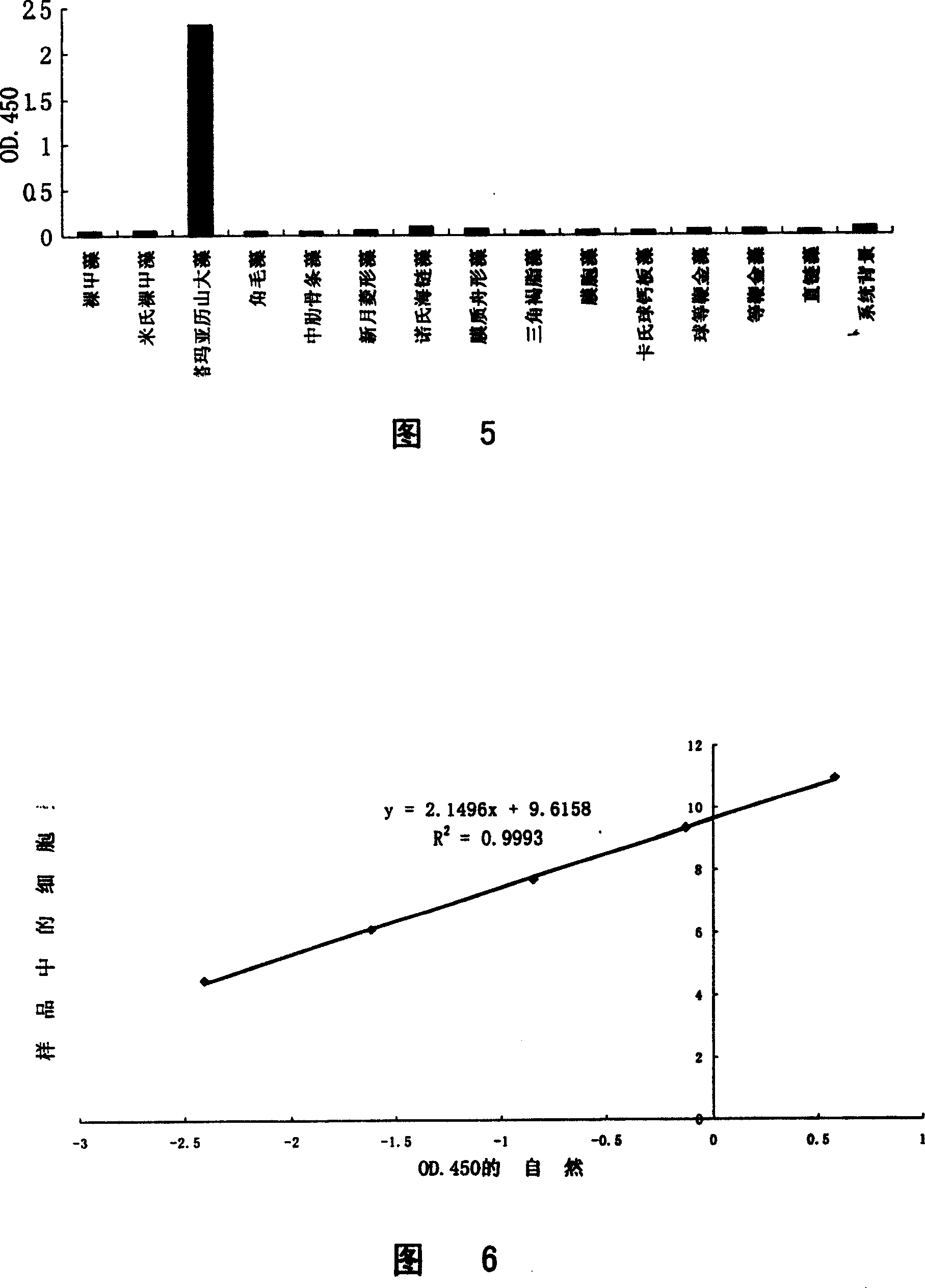
[0073] By determining the 18S rRNA sequence of the ribosomal small subunit of Chaetoceros algae and performing multiple sequence comparisons with the rRNA sequences of other algae, it was found that the 550-700 region of the small subunit rRNA was different from other algae, and the sequence of this region was determined to be a characteristic sequence; The S1 enzyme protection analysis probe CHV_S1 was designed and synthesized according to th...
Embodiment 2
[0104] Example 2: Qualitative and quantitative analysis of Alexandrium tamarensis by using S1 enzyme protection analysis and sandwich hybridization technology for rRNA
[0105] In this embodiment, the S1 enzyme protection analysis technique and the multiple sandwich hybridization technique are combined to realize the qualitative and quantitative analysis of Alexandrium tamarense, and the steps are as follows:
[0106] 2.1 The search for the specific region of Alexandrium tamarina rRNA and the design and synthesis of the S1 enzyme protection analysis probe
[0107] By determining the 18S rRNA sequence of the ribosomal small subunit of Alexandrium tamarica and performing multiple sequence comparisons with the rRNA sequences of other algae, it was found that the 600-800 region of the small subunit rRNA was different from other algae, and the sequence of this region was determined to be a characteristic sequence ; Design and synthesize the S1 enzyme protection analysis probe ALT S...
Embodiment 3
[0135] Example 3: Qualitative and quantitative analysis of Chaetoceros using S1 enzyme protection analysis for rRNA and sandwich hybridization technology
[0136] In this example, the steps of Example 1 were repeated, the only difference being that instead of using a universal signal probe, fluorescein was directly labeled at the 5' end of the specific signal probe (that is, connecting the probe and the signal probe The functions of the needle are combined into one).
[0137] As a result, the qualitative results of Chaetoceros completely the same as those in Example 1 were obtained, and the quantitative detection results were almost completely the same. However, the disadvantage of this embodiment is that a specific signal probe with a label needs to be synthesized for each group of detection probes, and the cost of synthesis is relatively high, while the general signal probes used in Examples 1 and 2 can be widely used. Universal, it is not necessary to synthesize a specific...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com