Method of GTP combining experimental signal window of enlarging GPCR tagging
A technology of isotope labeling and signal window, which is applied in the direction of material inspection products, preparation of test samples, biological testing, etc., can solve the problems of Saponin chemokine receptor effect, negative effect, GPCR no effect, etc. that have not been studied
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] Example 1 Establishment of CHO / CXCR1, CHO / CXCR4, and CHO / CCR5 cell lines
[0085] The target gene was cloned by RT-PCR from the total cDNA library of human leukocytes (source of whole blood: Shanghai Blood Center) to obtain full-length DNA encoding CXCR1, CXCR4 and CCR5 respectively (see below for the primers used to obtain the required DNA). Restriction endonuclease HindIII / XhoI (purchased from Promega) double-digested their full-length DNA and empty plasmid pcDNA3 (purchased from Invitrogen), and then inserted their full-length DNA into pcDNA3 by T4 ligase (purchased from Promega) , were obtained plasmid pcDNA3-CXCR1, pcDNA3-CXCR4 and pcDNA3-CCR5. The sequences of these plasmids were confirmed by sequencing.
[0086] CXCR1 PCR primers:
[0087] P1: 5'TCTAAGCTTACGCGGCGTT3' (SEQ ID NO: 1);
[0088] P2: 5'TACCTCGAGCCTGTCCAGAGA 3' (SEQ ID NO: 2).
[0089] CXCR4 PCR primers:
[0090] P1: 5'TCTAAGCTTAACTTCAGTTTG3' (SEQ ID NO: 3);
[0091] P2: 5'TACCTCGAGAATTCAAATTGATG...
Embodiment 235
[0098] Example 2 [ 35 S] GTPγS binding assay
[0099] 2.[ 35 The general procedure for S]GTPγS binding experiments is as follows:
[0100] ①Collect the corresponding cells obtained in Example 1 that were cultured for 48-72 hours, add cold lysate (5mM Tris-HCl, pH7.5, 5mM EDTA, and 5mM EGTA) and repeatedly pipette the cells 6-7 times with an insulin needle to make the cells crack. Centrifuge at 13000r / min at 4°C for 15 minutes, remove the supernatant and collect the cell membrane pellet.
[0101] ②According to the amount of cell membrane, use an appropriate volume of reaction solution (20mM HEPES, 100mM NaCl, and 5mM MgCl 2 , pH7.4) and resuspended with an insulin needle repeatedly for 6-7 times to form a uniform cell membrane suspension.
[0102] ③Establish the reaction system: the total volume is 100 μl, in which the cell membrane is 10 μg, [ 35 S] GTPγS (purchased from Amersham) 0.5nM (1,200Ci / mmol), GDP (purchased from Sigma) 40μM, then add agonists (both purchased fr...
Embodiment 3
[0108] Add Saponin in the reaction system of embodiment 3 [ 35 S] GTPγS binding assay
[0109] Using the CHO / CXCR1 prepared in Example 1, in step ③ of Example 2, add Saponin at a specified concentration in the reaction system (concentrations are respectively 5 μg / ml, 10 μg / ml, 20 μg / ml, 30 μg / ml, and IL-8 is used as an agonist. Other reaction conditions and operating steps are the same as in Example 2.
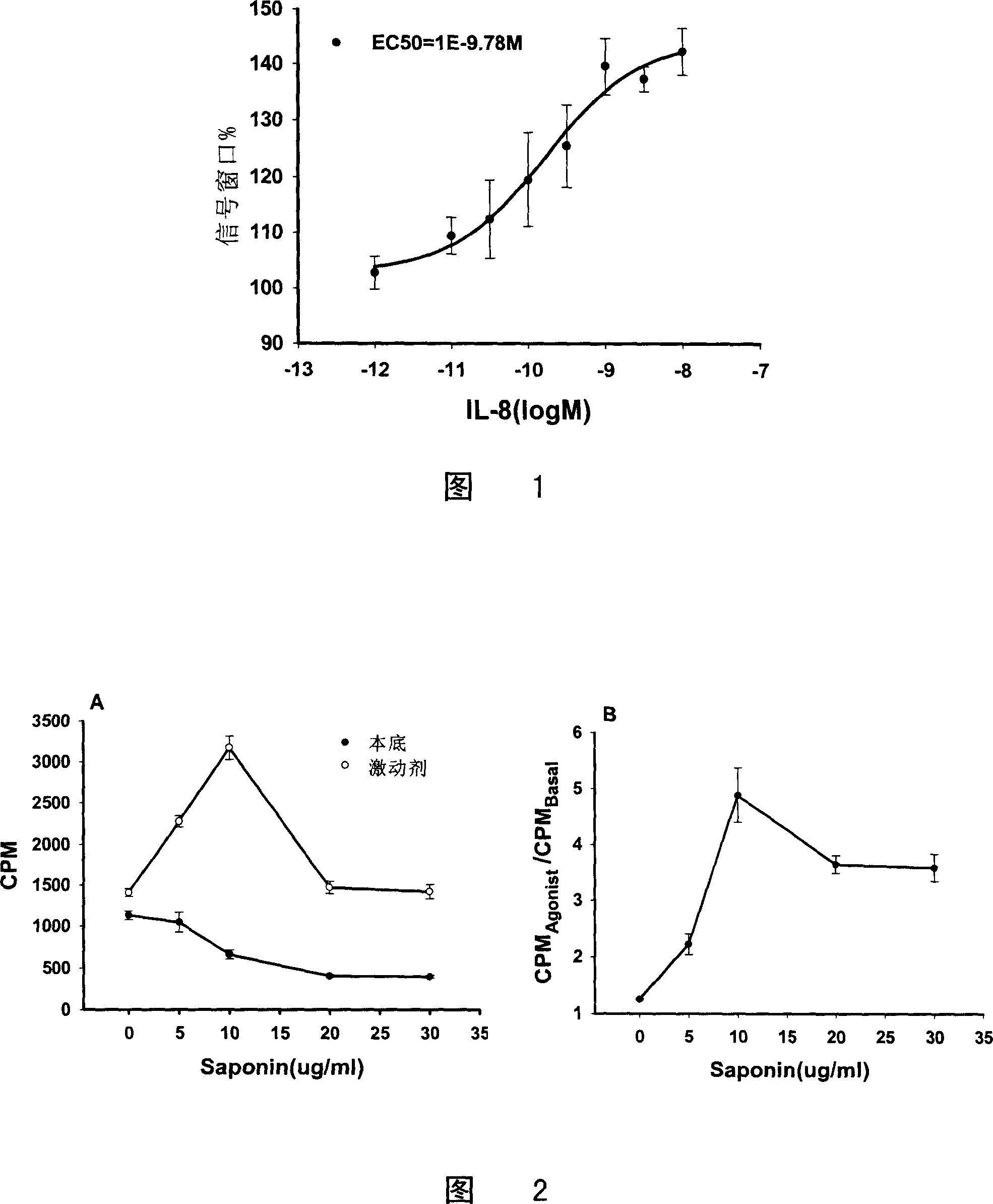
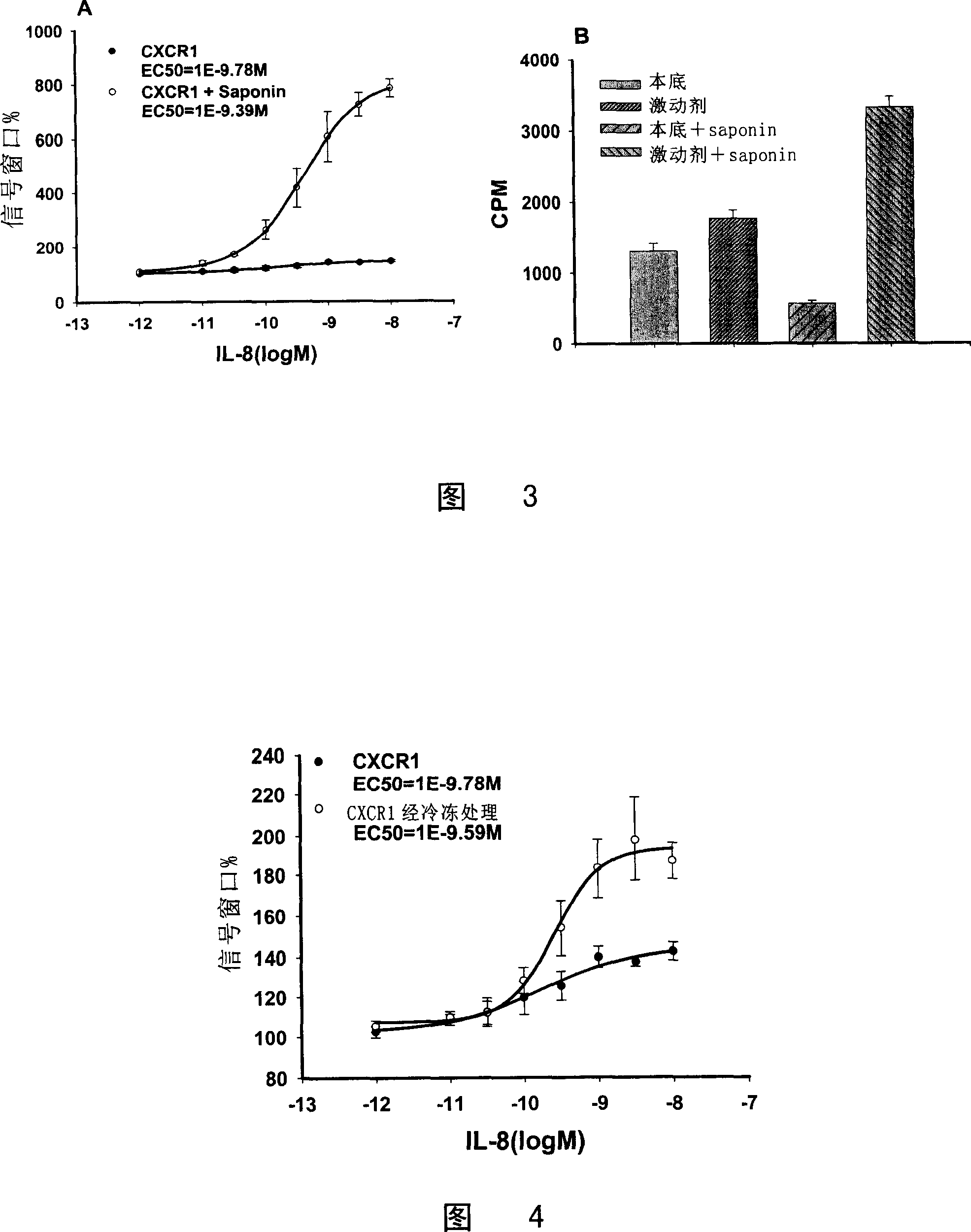
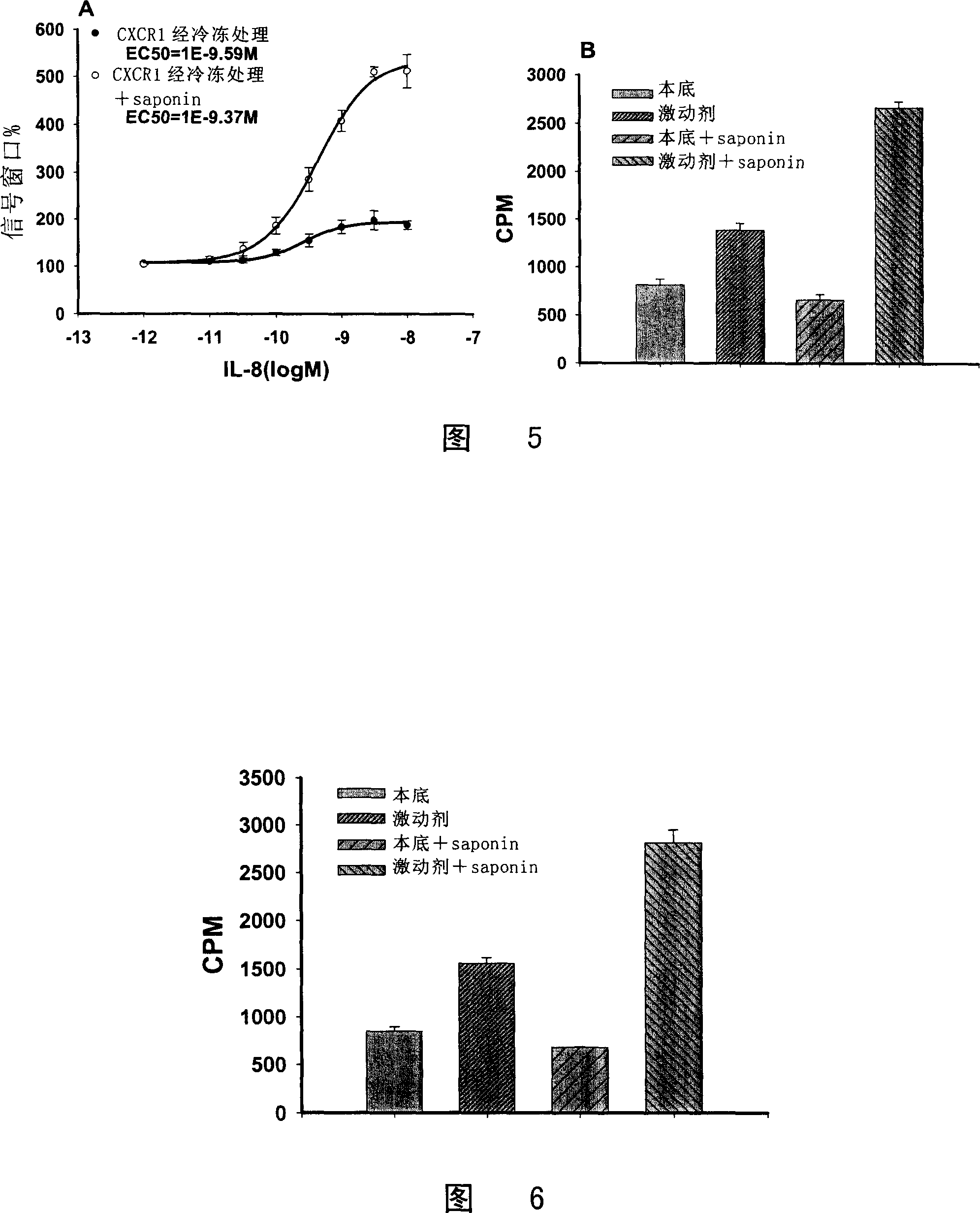
[0110] 1. CHO / CXCR1 [ 35 S] The relationship between the signal window of GTPγS binding experiment and the dosage of Saponin
[0111] The results showed that CPM Agonist The curve with the dose of Saponin is peak-shaped, and 10 μg / ml of Saponin can make CPM Agonist Maxed out, CPM Basal It gradually decreased with the increase of Saponin dose (Fig. 2A). The signal window reached its maximum at Saponin 10 μg / ml (Fig. 2B).
[0112] 2.10μg / ml Saponin can maximize the expansion of CHO / CXCR1[ 35 S] Signal window for GTPγS binding experiments
[0113] The results showed that a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com