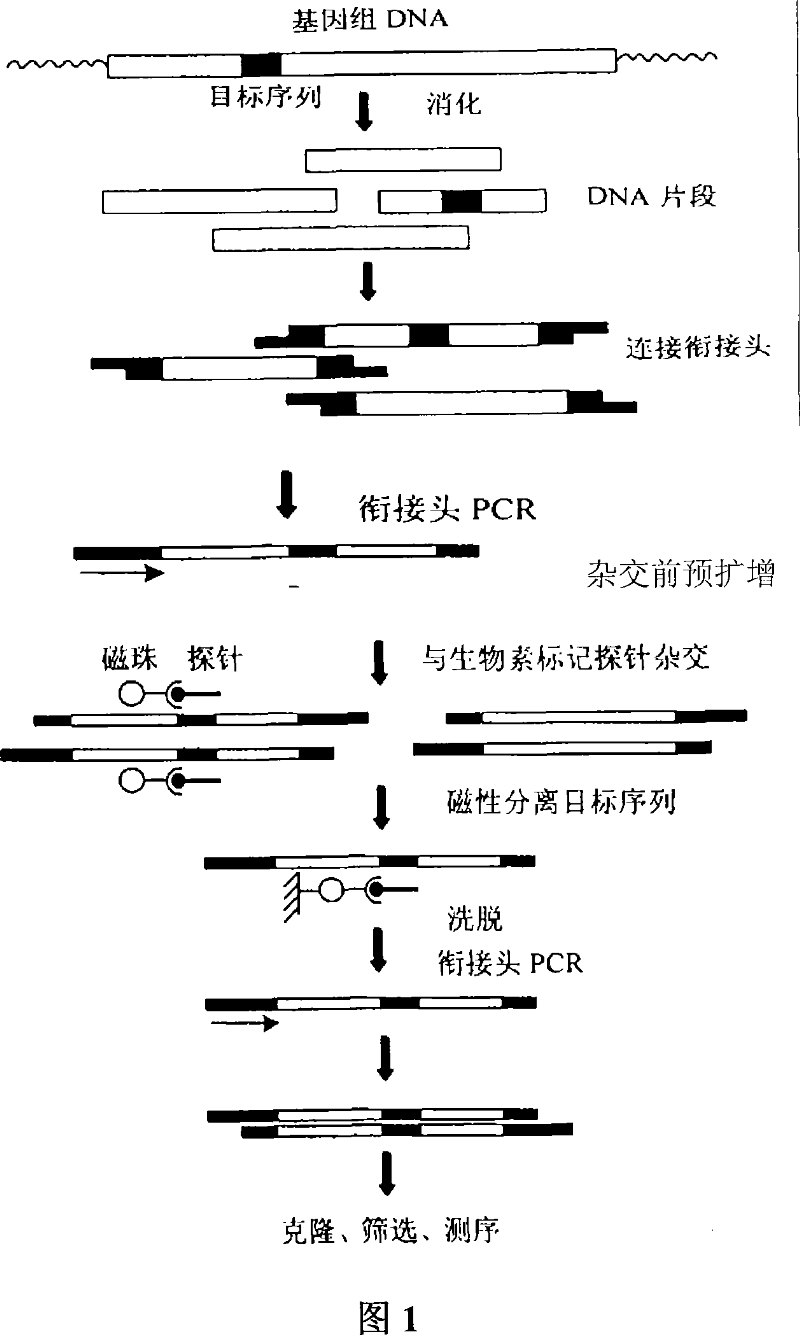
Construction method for Chlymys nobilis satellite mark
A technology of microsatellite marker and construction method, applied in the field of molecular biology DNA marker technology and application, can solve the problems of complicated steps and large workload, and achieve the effects of improving efficiency, simple operation and real and reliable results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0037] The following examples are further illustrations of the present invention and should not be regarded as limiting the present invention.
[0038] 1. Extraction of DNA
[0039] Take 100 mg of the adductor muscle of Chlamysgui scallop, shred it, place it in 0.7 ml of extraction buffer, add proteinase K (final concentration: 0.1 mg / ml), and digest overnight at 37°C. The reactant was extracted with phenol:chloroform (1:1), precipitated with isopropanol, and then dissolved in TE. DNA samples were treated with RNase (20 μg / ml) at 37° C. for 1 hour, and then purified with phenol / chloroform extraction solution. The samples were detected by electrophoresis, and the concentration was determined and stored at -20°C until use.
[0040] 2. Restriction ligation and pre-amplification before hybridization
[0041] Digest the genomic DNA of Chlamydia grandis with Sau 3AI, digest overnight at 37°C, the total volume is 100μl: Genomic DNA 10μg, 10×buffer, 10μl, 10u of DpnII, add water to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com