Method for rapid identification of liriomya huidobresis blanchard similar species by using PCR-RFLP technology
A technique of PCR-RFLP and Liriomyza, applied in the molecular field of identifying Liriomyza trifolii and its related species, can solve the problems of expensive endonucleases and inability to clearly distinguish them, and achieve the effect of low-cost and rapid detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
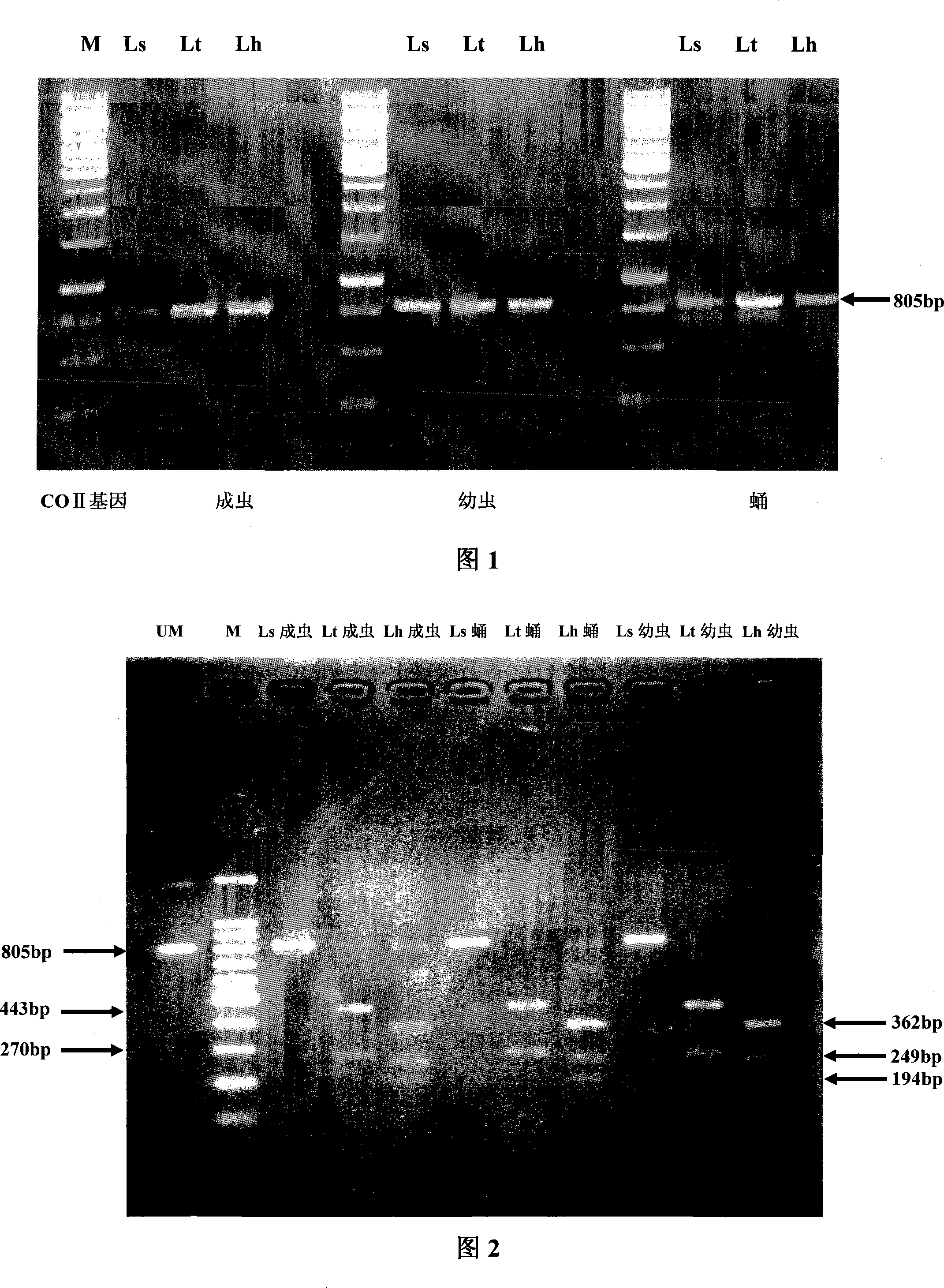
[0041] Example 1: (COII enzyme digestion identification of Liriomyza trifolii, Liriomyza sativae and Liriomyza sativae using DraI)
[0042] Follow these steps:
[0043] 1. DNA extraction and related sequence acquisition:
[0044] Take one adult specimen of Liriomyza trifoliata, Liriomyza sativae and Liriomyza sativae, and extract DNA respectively according to the following steps: Take one specimen and soak it in a 1.5ml centrifuge tube filled with sterile water for 1 hour; take out Grind the specimen in 100 μl extract (Tris: 10 mM, EDTA: 1 mM, SDS: 1%, PH=8.0), add 2.5 μl proteinase K (20 mg / ml) and digest in a water bath at 56 °C for 2 h; add an equal volume of phenol / Extract with chloroform / isoamyl alcohol (25:24:1) mixture, take the supernatant and add 2 times the volume of absolute ethanol to precipitate DNA; finally dissolve DNA with sterile water; store at -20°C as a template for PCR reaction .
[0045] The PCR reaction conditions are as follows: 50 μl of the system,...
Embodiment 2
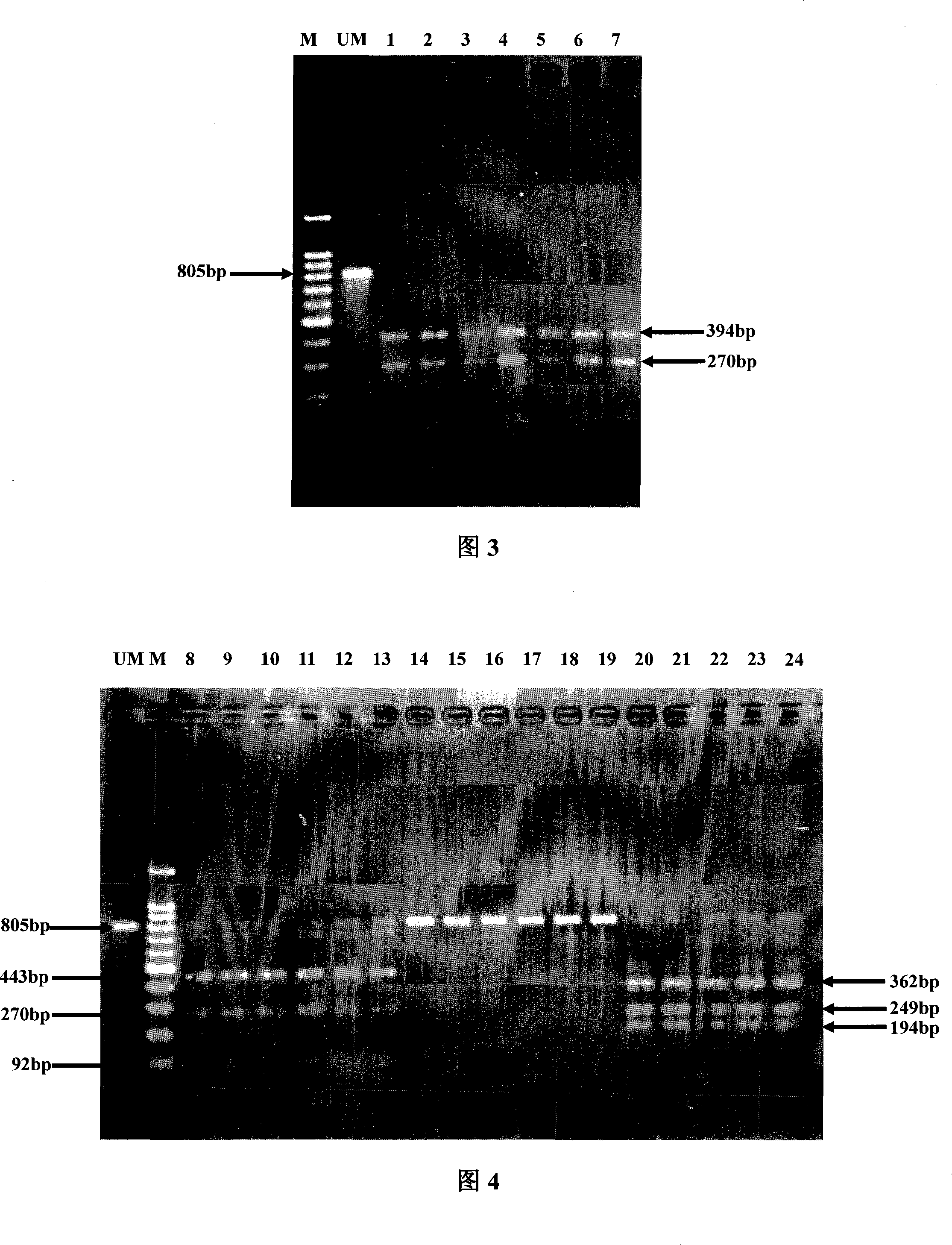
[0051] Embodiment 2: (Utilize DraI endonuclease to the verification of the homogeneity of different worm states of three kinds of Liriomyza sativae)
[0052] The extraction of DNA from pupae and larvae of Liriomyza trifolii, Liriomyza sativae and larvae of Liriomyza sativae and the acquisition of related sequences are the same as in Example 1; Adults were consistent, therefore, the discrimination analysis using DraI endonuclease will not be disturbed by insect state (Figure 2).
Embodiment 3
[0053] Embodiment 3: (Utilize DraI endonuclease to the verification of different species of Liriomyza pisa and three kinds of Liriomyza sativae)
[0054] Since Liriomyza pisa, which is similar in shape to the three types of Liriomyza sativae, overlaps with Liriomyza sativae on the host, it is necessary to use Liriomyza pisa as a verification species for verification; Same as Example 1), its electrophoresis bands are obviously different from three kinds of Liriomyza sativus enzyme-cut electrophoresis bands, indicating that the identification using DraI endonuclease will not be affected by mixed Liriomyza pisa (Fig. 3).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com